the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Network complexity of rubber plantations is lower than tropical forests for soil bacteria but not for fungi
Chuan Yang
Zhixiang Wu
Bangqian Chen
Xicai Zhang
Soil microbial communities play a crucial role in ecosystem functioning. Past research has examined the effects of forest conversion on soil microbial composition and diversity, but it remains unknown how networks within these communities respond to forest conversion, including when tropical rainforests are replaced with rubber plantations. Microbial networks are viewed as critical indicators of soil health and quality. They consist of two parts: nodes and edges. In this study, we used data from Illumina sequencing and shotgun metagenome sequencing to analyze bacterial and fungal community network structure in a large number of soil samples from tropical rainforests and rubber plantation sites on Hainan Island, China. Our results showed that only 5 %–10 % of shared network edges (i.e., links between species A and B existing in both rubber plantations and rainforests) were observed in both bacterial and fungal communities, which indicates that forest conversion altered the soil microbial network structure. The identity of keystone operational taxonomic units (OTUs) differed entirely between rubber plantation and rainforest sites, further underscoring the altered network structure. More edges and more negative correlations within the soil bacterial–fungal networks were observed at rubber plantation sites (dry season: 4284 total edges, 844 negative; rainy season: 7257 total edges, 1744 negative) than at rainforest sites (dry season: 3650 total edges, 149 negative; rainy season: 6018 total edges, 489 negative), demonstrating that soil bacterial–fungal network structure was more complex and stable in rubber plantations than in rainforests. For bacteria, a larger number of network edges were observed among bacterial networks in samples from tropical rainforest than in samples from rubber plantations, indicating that rainforest bacterial networks were more complex than those from rubber plantations. However, soil fungal networks from rubber plantations showed more links, suggesting that forest conversion increased fungal network complexity. More edges of network and more links between species and functions were observed in the rainy season than in the dry season, indicating that seasonal changes had a strong effect on network structure and function. Further analysis shows that soil pH, potassium (AK), and total nitrogen (TN) had more links with species of some phyla. In conclusion, forest conversion results in an increase in soil pH as well as a decrease in AK and TN, and these changes as well as seasonal variations had a great impact on soil microbial composition, network structure and function.
- Article
(7836 KB) - Full-text XML
-
Supplement
(1374 KB) - BibTeX
- EndNote
“Soil microbiome” refers to the enormous variety of microorganisms inhabiting the complex soil environment. It is highly diverse and comprises up to one-quarter of Earth's species (Wagg et al., 2019). Soil microbes play a critical role in the maintenance of soil quality and function, and they represent more than half of the biodiversity of terrestrial ecosystems (Philippot et al., 2013). A number of studies have investigated how the conversion of tropical forests to rubber plantations impacts soil microbial composition and diversity (Schneider et al., 2015; Kerfahi et al., 2016; Lan et al., 2017a, b, c, 2020a, b, 2021). Studies conducted in Indonesia (Schneider et al., 2015), Malaysia (Kerfahi et al., 2016) and southern China (Lan et al., 2017a, b, c) have found significant differences between rubber plantations and tropical forests, specifically that the diversity of soil bacteria was higher in rubber plantations than in rainforest. Compared with primary forests, agricultural systems tend to have higher bacterial richness but lower fungal richness (Lan et al., 2017a; Cai et al., 2018; Tripathi et al., 2012; Kerfahi et al., 2016). Compared with Eucalyptus plantations, rubber plantations have been found to possess a higher diversity of both bacteria and fungi (Ma et al., 2019). Song et al. (2019) reported that tropical forest conversion to rubber plantation results in reduced fungal microbial community network complexity. However, there are few studies on the impact of forest conversion on the soil bacterial community network structure nor on the drivers leading to changes in network structure.
The soil microbial network is viewed as a critical indicator of soil health and quality (Kuperman et al., 2014). Microbial networks consist of two parts: nodes and edges. Nodes indicate microbe types, generally operational taxonomic units (OTUs) or species, but can also indicate environmental variables of interest, such as soil pH. Edges (or links) indicate significant correlations between nodes. The number of links connected to a node is known as the node's degree (Röttjers and Faust, 2018). Usually, the more links a network has, the more complex the community is. Similarly, the higher the degree of a node (OTU or species), the more important the species is to the network structure (Berry and Widder, 2014). In recent years, microbial network analysis has been used to identify keystone taxa (Banerjee et al., 2018) and provide comprehensive insight into the microbial community structure and assembly (Fuhrman, 2009). The complexity of network structure is understood by calculating the number of edges, whereas keystone species of the community are found by estimating the degree of each species. The stability of the microbial community is determined by calculating the ratio of positive to negative correlations of the network, as a microbial community with a large proportion of positive correlations is thought to be unstable (Coyte et al., 2015).
Previous work has shown that agricultural intensification can reduce microbial network complexity (Banerjee et al., 2019). Logging alters the soil fungal network in tropical rainforests, i.e., more organized fungal communities in cut stands compared with primary stands (Chen et al., 2019). Soil bacterial networks are less stable under drought conditions than fungal networks (De Vries et al., 2018). While the exact drivers of microbial network structure still remain unknown, previous work has shown that soil nutrients, such as soil phosphorus content, and soil pH are the main drivers of the network structure for microbial communities (Banerjee et al., 2019). Seasonal variation also affects the network structure by changing the identity of keystone species in the community, given that a keystone species might be present only during a specific season or time period (Banerjee et al., 2018).
Hainan Island is home to a large area of tropical rainforest that is rich in biodiversity. It is a part of the Indian–Malay rainforest system at the northern edge of the world's rainforest distribution. However, rubber plantations now account for almost a quarter of the total extent of vegetated areas on Hainan Island (Lan et al., 2021). Previous work on the island has shown that forest conversion from rainforest to rubber plantations can result in shifts in bacterial composition, from Proteobacteria to Chloroflexi, and fungal composition, from Basidiomycota to Ascomycota (Lan et al., 2021). Other research has shown that microbial (including bacteria and fungi) diversity was higher in rainforest soils than in rubber plantations (Lan et al., 2021). Therefore, we hypothesize the following:
-
The conversion of rainforest to rubber plantations results in changes in the dominant phyla of soil microbes, with concomitant changes in the network structure and identity of keystone species.
-
Soil bacterial–fungal network structure is less complex but more stable in rubber plantations than in rainforest because high species diversity leads to complex network structure and unstable microbiome communities (Coyte et al., 2015).
-
Soil bacterial networks in rubber plantations are less complex than in rainforests because intensive cropping reduces the complexity of bacterial network, even as richness increases (Karimi et al., 2019).
-
Soil fungal network structure at rubber plantation sites is more complex than in the rainforest because organic farming has been shown to result in a much more complex fungal network than conventional or no-tillage farm systems, due to the use of organic fertilizer and the return of litter to the field (Banerjee et al., 2019).
By testing these hypotheses, we want to clarify the drivers and mechanisms of microbial community assembly that link forest conversion to differences in the soil microbial network structure. This study will provide critical information for understanding and managing microbial communities in the tropical forests of China and elsewhere.
2.1 Study site
This study was conducted on Hainan Island (18∘10′–20∘10′ N and 108∘37′–111∘03′ E), in southern China. The total area of Hainan Island is about 34 000 km2 (Lopez et al., 2009). It is the largest island within the Indo-Burma Biodiversity Hotspot of tropical Asia (Francisco-Ortega et al., 2010) and has a tropical monsoon climate. Hainan Island has a warm and humid climate year-round, with an average annual temperature of 22–26 ∘C. Rainfall is abundant, ranging from 1000 to 2600 mm yearly, with an average annual precipitation of 1639 mm. The rainy season occurs from May to October, with a total precipitation of about 1500 mm, accounting for 70 %–90 % of the total annual precipitation. Only 10 %–30 % of the total annual precipitation falls within the dry season, from November to April. The central part of Hainan Island is mountainous and contains old-growth tropical rainforests and monsoon forests. Rubber plantations are found on the plateaus surrounding the central mountainous zone.
2.2 Soil sampling
The tropical rainforests of Hainan are mainly distributed in Bangwangling, Diaoloshan, Wuzhishan, Yinggeling and Jianfengling national forest parks. Therefore, we selected a tropical rainforest at each of these sites for our study objectives. There are about 530 000 ha of rubber plantations on Hainan Island. Five rubber plantations were selected, one in Wanning, Ledong, Danzhou, Haikou and Qiongzhong, respectively. These five rubber plantations are located in the east, south, west, north and middle of Hainan, respectively (Fig. S1 in the Supplement). We selected 25- to 30-year-old rubber trees (i.e., mature rubber plantations) for our study objectives. Latex harvest and fertilizer application are common management practices in rubber plantations. Usually, compound fertilizer (1–1.5 kg per tree) and organic fertilizers (20–25 kg per tree) were applied once or twice a year. More information on the study sites is provided in Table S1 in the Supplement. Before soil sampling, we sterilized the soil drill with 75 % alcohol. After the removal of the litter layer using a 5 cm diameter steel drill, topsoil (0 to 20 cm) was collected, homogenized and passed through a 2 mm mesh sieve. For each site, 13 soil samples were collected; thus, a total of 130 samples were collected between the rubber plantation and tropical rainforest sites. Soil sampling was performed twice in 2018, once in January (dry season) and once in July (rainy season), resulting in a total of 260 soil samples (130 per forest type). Soil samples were divided into two parts. One was used to analyze soil water content, soil pH, total nitrogen (TN), total phosphorus (TP), total potassium (TK), nitrate nitrogen (NN), ammonium nitrogen (AN), available phosphorus (AP) and potassium (AK). Soil water content (%) was measured gravimetrically. Soil pH was measured in a soil : water suspension (1 : 2.5, ) using a pH meter. TN was determined using micro-Kjeldahl digestion followed by steam distillation. TP and TK were measured following digestion with NaOH. NN and AN were determined by steam distillation and indophenol-blue colorimetry, respectively. Soil samples were extracted with NaHCO3, and the extract was then used to measure available soil phosphorus AP via molybdate-blue colorimetry. To measure soil AK, soil was extracted with ammonium acetate, and the extract was then loaded onto an atomic absorption spectrometer with ascorbic acid as a reductant (Chen et al., 2019). Soil properties of the rubber plantation and rainforest sites are shown in Table S2. The other part of each soil sample was stored in an ultralow-temperature (−80 ∘C) refrigerator for later DNA extraction.
2.3 DNA extraction and polymerase chain reaction (PCR) amplification
Microbial DNA was extracted from 0.5 g of soil using E.Z.N.A.® soil DNA kits (Omega Bio-tek, Norcross, GA, USA) following the manufacturer's protocol. The fungal ITS1 hypervariable region was amplified using the PCR primers ITS1F (5′-CTTGGTCATTTAGAGGAAGTAA-3′) and ITS2R (5′-GCTGCGTTCTTCATCGATGC-3′) (Adams et al., 2013). For bacteria and archaea, the V4 hypervariable region of the bacterial 16S rRNA gene was amplified using the PCR primers 515FmodF (5′-GTGYCAGCMGCCGCGGTAA-3′) and 806RmodR (5′-GGACTACNVGGGTWTCTAAT-3′) (Walters et al., 2016; Sampson et al., 2016). The PCR reactions were conducted using the following approach: an initial 3 min denaturation at 95 ∘C, 27 cycles of 30 s at 95 ∘C, 30 s of annealing at 55 ∘C, 45 s of elongation at 72 ∘C, and a 10 min final extension at 72 ∘ C.
2.4 Illumina MiSeq sequencing
Amplicons were extracted from 2 % agarose gels, purified using the AxyPrep DNA gel extraction kit (Axygen Biosciences, Union City, CA, USA) and quantified using a QuantiFluor™ single-tube fluorometer (Promega, USA). Purified amplicons were pooled in an equimolar solution and then sequenced (paired-end, 2×250 bp) on an Illumina MiSeq platform according to standard protocols.
Shotgun metagenomic sequencing libraries were prepared and then sequenced by Majorbio, Inc. (Shanghai, China) using the Illumina HiSeq 2000 platform. The NRGene gene catalog was aligned against the Kyoto Encyclopedia of Genes and Genomes (KEGG) database using BLAST (Basic Local Alignment Search Tool, version 2.2.28+) (Qin et al., 2010).
2.5 Bioinformatics and data analysis
Raw FASTQ files were demultiplexed and quality filtered using QIIME (Quantitative Insights Into Microbial Ecology, version 1.17; Caporaso et al., 2010). Operational taxonomic units (OTUs) were clustered with a 97 % similarity cutoff using UPARSE (Edgar, 2013), and chimeric sequences were identified and removed using UCHIME. Using the RDP (Ribosomal Database Project) classifier, the phylogenetic affiliation of each 16S rRNA gene and internal transcribed spacer (ITS) gene sequence was determined using a confidence threshold of 70 % with the SILVA 16S rRNA database and UNITE database, respectively (Amato et al., 2013). For each site, the relative abundance of different taxa (Good, 1953) and the Shannon diversity index were calculated based on resampled sequence data using mothur (Schloss et al., 2009). The raw reads were deposited into the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database (Accession Number: SRP108394, SRP278296, SRP278319).
2.6 Statistical analysis
To make the co-occurrence network analyses simple, we only used the top 300 most abundant bacterial and fungal OTUs to analyze the network structure of the two forest types. These OTUs roughly match the OTUs with a relative abundance greater than 0.05 % (Jiao et al., 2016). Four networks, representing each habitat during each sampling period (rainforest and rubber plantations in the dry and rainy seasons) were constructed with 65 samples each. We also performed the bacterial–fungal community network analysis to investigate the soil microbial network complexity of each of the four. Interactions consisted of Spearman's rank correlations. Co-occurrence networks were constructed using only significant correlations of ρ>0.6 (P<0.01) (Barberan et al., 2012), as this cutoff includes a range of interaction strengths (De Vries et al., 2018). The networks were then visualized in R using the “igraph” package. To reveal the distribution pattern of correlation coefficients, the frequency of the coefficients of ρ>0.3 (P<0.01) were plotted. The “NetworkAnalyzer” tool in Cytoscape (version 3.4.0) was used to calculate network topology parameters including number of nodes, edges, degree, betweenness and closeness. The number of shared edges and unique edges as well as keystone OTUs were calculated to evaluate whether the network structure changed. We defined a shared network edge as the link (edge) between two species (species A and species B) that not only exist in the rubber plantation network but also exist in the rainforest network. In contrast, a unique edge was defined as a link that only exists in either the rubber plantations or rainforests. The numbers of shared edges and unique edges were calculated using igraph and were visualized using Venn diagrams. Keystone OTUs are known to be important for ecosystem structure and function and were selected on the basis of high network degree, high closeness centrality, and low betweenness centrality, as defined by Berry and Widder (2014). To evaluate the proportional influence of each phylum on bacterial and fungal network structure, node degrees of each phylum were calculated and bar plots were created. Correlation coefficients between species and functions were calculated based on metagenomics data. Here, we used the top 50 most abundant species and the top 50 KEGG functions (pathway level 3). The species and function correlation network was then constructed on the Majorbio cloud platform (https://cloud.majorbio.com/, last access: 23 January 2022). To reveal the relationship between microbial taxa and environmental variables, two-way correlation networks were also constructed on the Majorbio cloud platform. The topological role of each node in a network was assessed by the Zi and Pi values, where Zi represents the connectivity of node i within a module, and Pi measures the degree of the connection of node i with other modules (Roger and Amaral, 2005). All species can be divided into four groups according to simplified criteria (Olesen et al., 2007), namely peripherals (Zi<2.5 and Pi<0.62), connectors (Pi>0.62), module hubs (Zi>2.5) and network hubs (Zi>2.5 and Pi>0.62). The Zi and Pi values were calculated using the “GIANT” package in Cytoscape. The Zi–Pi plot was created with “ggplot2” in R.
3.1 Bacterial and fungal networks
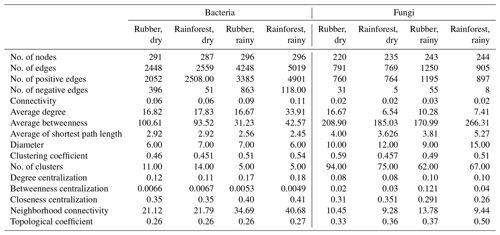
Our results showed that the bacterial–fungal community network was more complex in rubber plantations (4284 edges in the dry season, 7257 in the rainy season) than in primary forests (3650 in the dry season, 6507 in the rainy season) and that it was more complex during the rainy season than during the dry season (Fig. 1). Most nodes of bacterial networks (Fig. 2) and fungal networks (Fig. 3) varied with forest type in both the dry and rainy seasons. For bacterial networks, there were 2559 and 2501 edges in the forests and rubber plantations, respectively, in the dry season, but these two networks only shared 262 edges, accounting for only about 10 % the total edges (Fig. 4a, b). Similarly, these networks only shared 519 edges during the rainy season. For fungal networks, there were only four and five shared edges (i.e., no more than 5 % of the total edges) in the dry and rainy seasons, respectively (Fig. 4c, d).
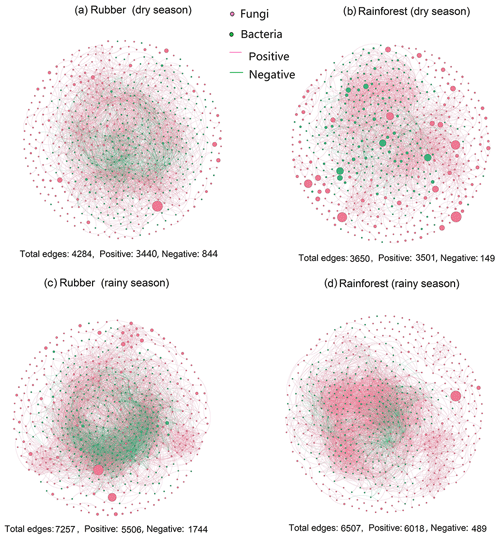
Figure 1The soil bacterial–fungal community network of rubber plantations and rainforests in the dry and rainy seasons. Red solid circles indicate fungi, and green solid circles indicate bacteria. Red lines indicate a positive correlation between OTUs, and green lines indicate a negative correlation. The absolute value of the correlation coefficient ρ>0.6 (P<0.01).
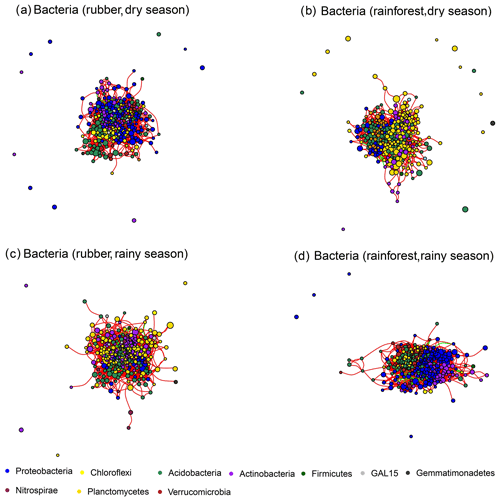
Figure 2The soil bacterial network structure of rubber plantations and tropical rainforests in the dry and rainy seasons. Red lines indicate a positive correlation between OTUs, and green lines indicate a negative correlation. The absolute value of the correlation coefficient ρ>0.6 (P<0.01).
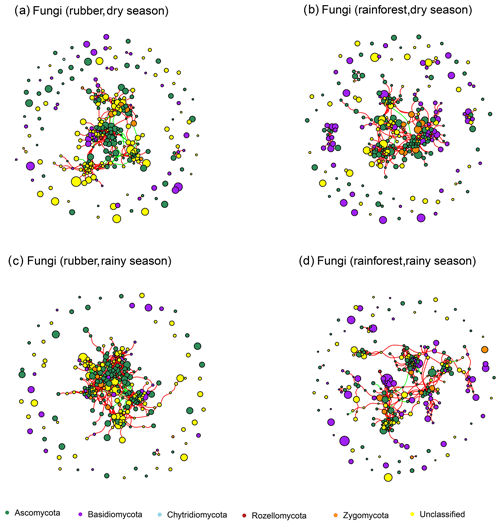
Figure 3The soil fungal network structure of rubber plantations and tropical rainforests in the dry and rainy seasons. Red lines indicate a positive correlation between OTUs, and green lines indicate a negative correlation. The absolute value of the correlation coefficient ρ>0.6 (P<0.01).
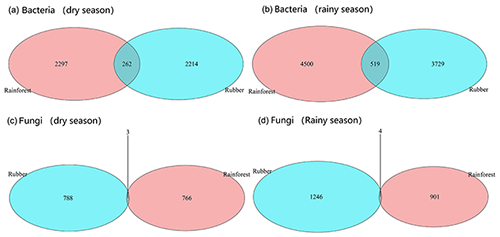
Figure 4The number of shared and unique edges of soil bacterial and fungal networks in rubber plantations and tropical rainforests in the dry and rainy seasons. The number where the two circles cross is the number of shared edges. Numbers in the red circles present the unique edges in rainforests, whereas numbers in blue circles present the unique edges in rubber plantations.
The number of edges of bacterial and fungal networks were almost equivalent during the dry season. However, in the rainy season, there were more edges in the rainforest bacterial network than in the rubber plantation network (Table 1, Fig. 4b). For the network structure of the fungal community, more edges were observed in rubber plantations in the rainy season (Table 1, Fig. 4d). Similarly, there were no significant differences in the degree of the bacterial or fungal networks between the tropical rainforest sites and rubber plantations in the dry season (Fig. 5a, c). In the rainy season, rainforest sites had a higher bacterial network degree, while rubber plantations showed a higher fungal network degree (Fig. 5b, d). For bacterial networks, more nodes (OTUs) with a high degree (rubber plantation had two nodes with a degree greater than 75, rainforest had eight such nodes) were observed in the rainforest during the rainy season (Fig. S2B). For fungal networks, 15 nodes of a higher degree (degree greater than 25) were observed in rubber plantations; however, only 7 such nodes existed for rainforest sites (Fig. S2D). These results indicate that the fungal network structure in the rubber plantations was more complex than for tropical rainforests during the rainy season but that the reverse was true for bacteria.
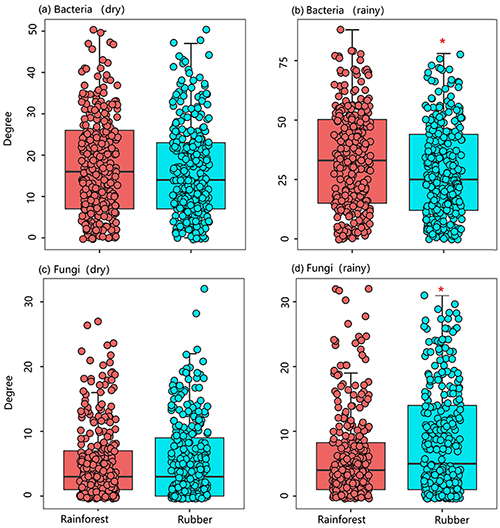
Figure 5The network degree of soil bacterial and fungal communities of rubber plantations (blue) and tropical rainforests (red) in the dry and rainy seasons.
Table 1Topological properties of the soil microbial (bacterial and fungi) network structure in a rubber plantation and tropical rainforest in the dry and rainy seasons.
When considering the ratio of positive to negative correlation coefficients, the bacterial–fungal community network revealed that rubber plantations (844 edges in the dry season, 1744 edges in the rainy season) have more negative links than rainforests (149 edges in the dry season, 489 edges in the rainy season), indicating that the rubber plantation network was more stable than that of the rainforests (Fig. 1). More correlations (greater than 0.3, P<0.01) were also calculated to investigate the positive to negative correlation ratio, and the results showed that negative correlations between bacterial and fungal OTUs in the rubber plantations were consistently stronger than in the tropical rainforests in both the dry season and the rainy season (Fig. 6).
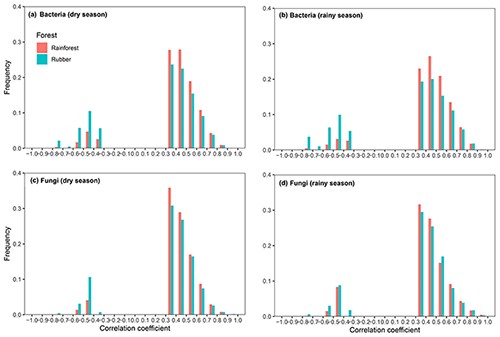
Figure 6Frequency distributions of the correlation strength in bacterial (a – dry season, b – rainy season) and fungal (c – dry season, d – rainy season) networks of rubber plantations and tropical rainforests in the dry and rainy seasons (absolute correlation coefficient greater than 0.3, P<0.01). Correlations in rainforest networks are red, and correlations in rubber plantation networks are blue.
3.2 Keystone taxa
Module hubs and network hubs have been commonly identified as keystone taxa in networks. However, for both the bacterial and fungal communities, neither tropical rainforest nor rubber plantation networks possessed module hubs and network hubs (Figs. S3, S4). The keystone taxa of bacteria were very different between rubber plantations and tropical rainforest sites in both the dry and rainy seasons. For bacteria, there were more keystone taxa at tropical rainforest sites than in the rubber plantations in both the dry and rainy seasons, indicating that the tropical rainforest networks had higher complexity. We found that some groups of Acidobacteria are keystone taxa at tropical rainforest sites but disappeared after forest conversion. There were more Actinobacteria in rubber plantations than at tropical rainforest sites (Table S3).
For fungi, more keystone taxa were observed in rubber plantations than at tropical rainforest sites during both the dry and rainy seasons, indicating that the rubber plantation networks were more complex. Most keystone taxa belong to Ascomycota, suggesting that members of this group are very important for network structure. In addition to forest conversion, seasonal changes also affect the keystone taxa of the fungal community network. There were more Basidiomycota OTUs in the dry season but more Ascomycota in the rainy season (Table S4).
3.3 Drivers of microbial composition and network structure
Two-way correlation network analysis of the 300 most abundant OTUs and environmental variables revealed that elevation, AK and soil pH were the main drivers of bacterial community composition and had 106, 104 and 86 degrees, respectively (Fig. S5A). For fungi, elevation had the highest degree (61), followed by AK (51) (Fig. S5B). The relationship between OTU abundance and soil pH revealed that the soil pH was negatively correlated with the abundance of some species of Acidobacteria but was positively correlated with the abundance of some species of Chloroflexi and some species of Ascomycota (Fig. S6). AK was positively correlated with the abundance of Planctomycetes, Verrucomicrobia and Basidiomycota species but was negatively correlated with the abundance of Chloroflexi and Ascomycota. In other words, AK and soil pH were important drivers of microbial composition.
The total degree of each phylum was used to reveal the influence of each phylum on network structure (Fig. 7). For bacteria, Proteobacteria, Actinobacteria and Acidobacteria had a large influence on network structure. Acidobacteria and Planctomycetes contributed more to rainforest networks than to rubber plantation networks. However, Actinobacteria and Chloroflexi showed the opposite pattern. For fungi, Ascomycota and Basidiomycota had large influences on network structure. Both Ascomycota and Basidiomycota had stronger influences on rainforest networks than rubber plantation networks. The influence of Ascomycota was stronger during the rainy season than during the dry season, indicating that seasonal change also had an impact on fungal community networks. The degree of each phylum of bacterial and fungal network also revealed that seasonality had effects on the network degree (Fig. S7).
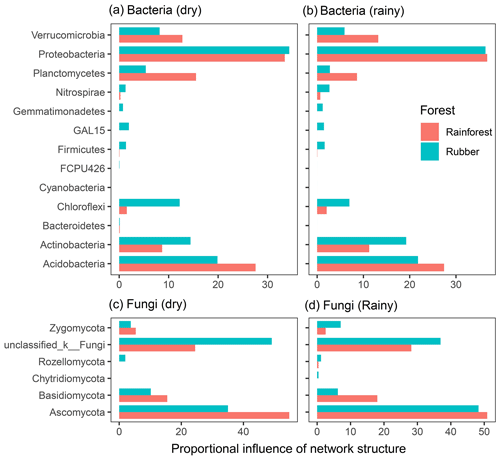
Figure 7The proportional influence of different phyla on the bacterial and fungal network structure in both the dry and rainy seasons. The influence was the number of degrees of nodes belonging to a particular phylum: panel (a) shows bacteria in the dry season, panel (b) shows bacteria in the rainy season, panel (c) shows fungus in the dry season and panel (d) shows fungus in the rainy season.
3.4 Interaction between species and functions
Two-way network analysis of the 50 most abundant species (all belonging to bacteria groups based on metagenomic data) and the 50 most abundant KEGG functions revealed the interaction between microbial composition and functions. The results showed that soil network structure at rainforests sites (460 edges in the dry season, 1750 in the rainy season) was more complex than that of rubber plantations (223 in the dry season, 451 in the rainy season) (Fig. 8), indicating that more links between microbial species and functions were observed in the soils of rainforest sites. Both rubber plantation and rainforest networks were more complex in the rainy season than in the dry season. We also found that metabolism was the most important function in soil microbial networks. Surprisingly, species of Actinobacteria were negatively correlated with function in rubber plantations.
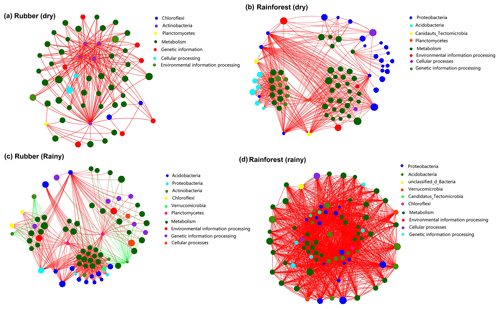
Figure 8Network of the top 50 most abundant species (based on metagenomics data) and the top 50 most frequent KEGG functions (pathway level 3) of rubber plantation and tropical rainforest sites in the dry and rainy seasons: panel (a) shows rubber in the dry season, panel (b) shows rainforest in the dry season, panel (c) shows rubber in the rainy season and panel (d) shows rainforest in the rainy season. The size of the node indicates the species/function abundance. A red line indicates a positive correlation between species/functions, and a green line indicates a negative correlation. The absolute value of the correlation coefficient >0.6 (P<0.01).
4.1 Forest conversion reduces soil bacterial network complexity
Land-use changes increasingly threaten biodiversity, particularly in tropical forests (Gibson et al., 2011). However, we still have little understanding of how soil networks respond to forest conversion, such as when rainforests are converted to rubber plantations. Our results showed that forest conversion had large effects on both soil bacterial and fungal networks. Tropical rainforest bacterial networks had more edges (Table 1) and a higher degree (Fig. 4) than rubber plantation networks, especially during the rainy season, indicating that the rainforest bacterial network was more complex than the rubber plantation network. This finding is consistent with previous observations that soil bacterial networks were more complex in natural systems than in crop soil (Karimi et al., 2019). Further study showed that soil networks become more connected through the process of ecological restoration (Morriën et al., 2017). The observed decreases in network complexity and cohesion support the hypothesis that cropping may enhance the isolation of bacterial taxa (Karimi et al., 2019), which results in lower numbers of connections in the network. In addition, at the microscale, the structure of tilled soils is more homogeneous, and the soil pores are less connected than in soils that are not tilled (Pagliai et al., 2004), such as rainforest soil. In nature, soil ecosystems are highly heterogeneous, as hot spots of soil microbial biodiversity can form spatial and temporally within soil aggregates (Bach et al., 2018). This spatial heterogeneity likely plays an important role in the interactions among microbes and the mechanisms by which more complex and diverse communities drive various nutrient-cycling processes on small spatial scales (Wagg et al., 2019).
A large number of studies employing microbial network analysis have enriched our understanding of microbial co-occurrence patterns in various soil ecosystems. However, very little is known about whether differences in the structure of microbial networks have consequences for microbiome functioning (Wagg et al., 2019). Our results demonstrated that more species were correlated with metabolism in a natural system than in an agricultural system, especially during the rainy season. This is in line with a previous study conducted in Sumatra, Indonesia, which found that the transformation of forest to rubber plantation results in a 10 %–16 % decrease in community metabolism (Barnes et al., 2014). Fewer interactions between microbial species (most of them are bacteria) and functions in rubber plantations demonstrated that forest conversion reduced the soil bacterial network complexity.
4.2 Forest conversion increased soil fungal network complexity
Surprisingly, rainforest bacterial networks were characterized by fewer edges (Table 1) and a lower degree (Fig. 4) than the rainforest bacterial networks, meaning that rubber plantation bacterial networks were more complex than those of native forests. Our results were not consistent with a study conducted in Xishuangbanna (Song et al., 2019) which showed that tropical forest conversion reduced fungal network complexity, but out work is consistent with previous observations which found that fungal community networks were more organized in disturbed forest compared with primary forest (Chen et al., 2019). The observations of Banerjee et al. (2019) showed that organic agricultural fields harbored much more complex fungal networks with many more keystone taxa than conventionally managed fields. Forest conversion resulted in shifts in fungal composition from Basidiomycota to Ascomycota (Fig. S8), as seen in previous investigations (Lan et al., 2021, 2020a). Previous work showed that Basidiomycota species display higher drought sensitivity than Ascomycota species (Taniguchi et al., 2018); this would result in a shift in the richness and abundance of Basidiomycota species (Fig. S8). Many Basidiomycota species are capable of long-distance dispersal (Egidi et al., 2019; Geml et al., 2012), which may result in a decrease in fungal network complexity. This possibly explained why Ascomycota OTUs contribute more to the network structure than Basidiomycota (Fig. 6). Overall, reductions in the abundance and richness of Basidiomycota species led to an increase in fungal links in rubber plantations.
4.3 Forest conversion enhanced the stability of the soil network
The ability of a microbial community to resist and recover from disturbances is defined as the stability of the community (De Vries and Shade, 2013). The ratio of positive to negative network links constitutes the balance between facilitative and inhibitive relationships within a network (Karimi et al., 2017). Theoretical studies, for example, predict that ecological networks that consist of weak interactions are more stable than those with strong interactions (Neutel et al., 2002; Coyte et al., 2015) and that compartmentalization and the presence of negative interactions increase the stability of networks under disturbances (Coyte et al., 2015; Rooney et al., 2006; Stouffer and Bascompte, 2011). In our study, more negative correlations were detected in the rubber plantation, indicating that the network structure of rubber plantation soils was more stable than that of rainforest soils (De Vries et al., 2018). Forest conversion from rainforest to rubber plantations resulted in shifts in bacterial composition from Proteobacteria to Chloroflexi and shifts in fungal composition from Basidiomycota to Ascomycota (Lan et al., 2021). Proteobacteria and Basidiomycota are copiotrophic microorganisms, whereas Chloroflexi and Ascomycota are oligotrophic (Yang et al., 2019; Lan et al., 2021). This indicated that forest conversion resulted in shifts in soil microbial composition from more k-strategists to more r-strategists (De Vries and Shade, 2013). r-strategists are quick-growing and less resistant but are more resilient to disturbances. However, growth is not the only mechanism that could maintain community stability, as the responses of microorganisms to disturbance are very complex.
4.4 Main drivers of soil microbial composition and network structure
Forest conversion results in a loss of plant diversity, a loss of plant biomass and an increase in soil pH (Lan et al., 2017a, b). Rubber plantations had a significantly higher pH, which explains the relative decrease in the abundance of Acidobacteria (Lan et al., 2017a). Our results demonstrate that forest conversion drives a shift in which soil microbes act as keystone species (Table 1). We found that many OTUs of Acidobacteria fit our criteria as keystone species for rainforest sites, which is consistent with previous findings (Banerjee et al., 2018). Unexpectedly, OTU11388 and OTU11373, both Acidobacteria, were observed in rainforest soils in both the dry and rainy seasons, indicating that Acidobacteria were very important for rainforest soil bacterial networks (Fig. 6, Table S3). Higher AK concentrations resulted in a higher abundance and more taxa of Actinobacteria (Fig. 9), which suggests that Actinobacteria contributed more in rubber plantation than in rainforest (Fig. 6). Indeed, forest conversion reduced the abundance of Actinobacteria OTUs (Fig. S9). Due to human disturbance in rubber plantations, the soil will inevitably be slightly polluted with herbicides and domestic garbage. A previous study observed a member of Actinobacteria in contaminated soil (Jiao et al., 2016).
Forest conversion also increases land-use intensity (Brinkmann et al., 2019), including the application of fertilizer and herbicide. The application of herbicides has also been linked to significant decreases in root colonization and spore biomass of arbuscular mycorrhizal fungi in tropical agriculture (Zaller et al., 2014). Soil nutrient concentrations show a decline around the roots of rubber plantations compared with those from rainforests (Sahner et al., 2015). Our study was no exception. For instance, the AK and TN concentrations were significantly lower in rubber plantations than in samples from the rainforest sites (Table S2). Higher concentrations of AK reasonably explained the higher contribution of Basidiomycota to the network structure (Fig. 8b) due to the positive association of AK with Basidiomycota.
Spatiotemporal heterogeneity can be a major driver of the abundance and distribution of keystone taxa in soil, which is a highly heterogeneous and multifaceted environment (Mills et al., 1993; Power et al., 1996; Mouquet et al., 2013). Seasonal variability determines the structural and compositional properties of microbiomes in an environment; as such, a keystone species might be present only during a specific season or time period (Banerjee et al., 2018). It was interesting that more bacterial OTUs were identified as connectors during the rainy season than during the dry season. Connectors have been characterized as generalists (Olesen et al., 2007), and generalists drive covariation among communities in a network (Chen et al., 2019). Previous observations have demonstrated that some keystone taxa that were found in the dry season disappeared during the rainy season (Lan et al., 2018). Seasonal changes possibly explain why some keystone taxa were observed in the rainy season but not in the dry season.
4.5 Possible impact of forest conversion on microbial community function
Forest conversion results in a decrease in the abundance of Proteobacteria and an increase in Actinobacteria. Most species of Proteobacteria were positively correlated with metabolic function, whereas most of Actinobacteria were negatively associated with metabolic function (Fig. 8). Therefore, the changes in the abundance of these two phyla result in a reduction in microbial community function after forest conversion. Because the metabolic function of a specific species is often affected by environmental conditions (Louca et al., 2018), some species are not correlated with any function in the dry season but are correlated with metabolic function in the rainy season, indicating that some microorganisms do not participate in the metabolic process during the dry season, especially in the rubber plantations. In conclusion, forest conversion as well as seasonal variation had a great impact on soil microbial community functions.
Our knowledge about land-use impacts on soil ecosystems is mostly limited to biodiversity and ecosystem functions, leaving uncertainty about how soil networks change after forest conversion. This study is the most comprehensive report on changes in network structure that occur when tropical rainforests are converted into rubber plantations. Our study showed that forest conversion altered both bacterial and fungal soil networks, reduced bacterial network complexity and enhanced fungal network complexity, especially during the rainy season. One possible reason maybe that forest conversion altered soil pH and other soil properties, which altered bacterial composition, subsequent network structure and certain soil ecosystem functions. Our study demonstrates the impact of forest conversion on soil network structure, which has important implications for ecosystem functioning, such as metabolic activity, and the health of soil ecosystems in tropical regions.
The code and data generated in this study are available from the corresponding authors upon reasonable request.
The supplement related to this article is available online at: https://doi.org/10.5194/soil-8-149-2022-supplement.
GL was responsible for conceptualizing the study; developing the methodology; and writing, reviewing and editing the paper. CY, RS, BC, XZ and ZW carried out the investigation.
The contact author has declared that neither they nor their co-authors have any competing interests.
Publisher's note: Copernicus Publications remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
We appreciate J. Aaron Hogan and another anonymous reviewer very much for their positive and constructive comments and suggestions on our manuscript.
This work was supported by the National Natural Science Foundation of China (grant no. 31770661), the High-level Talents Project of Hainan Natural Science Foundation (grant no. 320RC733), the Finance Science and Technology Project of Hainan Province (grant no. ZDYF2019145) and the Earmarked Fund for China Agriculture Research System (grant no. CARS-33-ZP3).
This paper was edited by Ember Morrissey and reviewed by J. Aaron Hogan and one anonymous referee.
Adams, R. I., Miletto, M., Taylor, J. W., and Bruns, T. D.: Dispersal in microbes: fungi in indoor air are dominated by outdoor air and show dispersal limitation at short distances, ISME J., 7, 1262–1273, 2013.
Amato, K. R., Yeoman, C. J., Kent, A., Carbonero, F., Righini, N., Estrada, A. E., Gaskins, H. R., Stumpf, R. M., Yildirim, S., Torralba, M., Gillis, M., Wilson, B. A., Nelson, K. E., White, B. A., and Leigh, S. R.: Habitat degradation impacts primate gastrointestinal microbiomes, ISME J., 7, 1344–1353, 2013.
Bach, E. M., Williams, R. J., Hargreaves, S. K., Yang, F., and Hofmockel, K. S.: Greatest soil microbial diversity found in micro-habitats, Soil Biol. Biochem., 118, 217–226, 2018.
Banerjee, S., Schlaeppi, K., and van der Heijden, M. G. A.: Keystone taxa as drivers of microbiome structure and functioning, Nat. Rev. Microbiol., 16, 567–576, 2018.
Banerjee, S., Walder, F., Büchi, L., Meyer, M., Held, A. Y., Gattinger, A., Keller, T., Charles, R., and van der Heijden, M. G. A.: Agricultural intensification reduces microbial network complexity and the abundance of keystone taxa in roots, ISME J., 13, 1722–1736, 2019.
Barberan, A., Bates, S. T., Casamayor, E. O., and Fierer, N.: Using network analysis to explore co-occurrence patterns in soil microbial communities, ISME J., 6, 343–351, 2012.
Barnes, A. D. Jochum, M., Mumme, S., Haneda, N. F., Farajallah, A., Widarto, T. H., and Brose, U.: Consequences of tropical land use for multitrophic biodiversity and ecosystem functioning, Nat. Commun., 5, 5351, https://doi.org/10.1038/ncomms6351, 2014.
Berry, D. and Widder, S.: Deciphering microbial interactions and detecting keystone species with co-occurrence networks, Front. Microbiol., 5, 1–14, 2014.
Brinkmann, N., Schneider, D., Sahner, J., Ballauff, J., Edy, N., Barus, H., Irawan, B., Budi, S. W., Qaim, M., Danie, R., and Polle, A.: Intensive tropical land use massively shifts soil fungal communities, Sci. Rep.-UK, 9, 3403, https://doi.org/10.1038/s41598-019-39829-4, 2019.
Cai, Z. Q., Zhang, Y. H., Yang, C., and Wang, S.: Land-use type strongly shapes community composition, but not always diversity of soil microbes in tropical China, Catena, 165, 369–380, 2018.
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., Fierer, N., Peña, A. G., Goodrich, J. K., and Gordon, J. I.: QIIME allows analysis of high-throughput community sequencing data, Nat. Methods, 7, 335–336, 2010.
Chen, J., Xu, H., He, D., Li, Y. D., Luo, T. S., Yang, H. G., and Lin, M. X.: Historical logging alters soil fungal community composition and network in a tropical rainforest, Forest Ecol. Manage., 433, 228–239, 2019.
Coyte, K. Z., Schluter, J., and Foster, K. R.: The ecology of the microbiome: networks, competition, and stability, Science, 350, 663–666, 2015.
De Vries, F. T. and Shade, A.: Controls on soil microbial community stability under climate change, Front. Microbiol., 4, 265, https://doi.org/10.3389/fmicb.2013.00265, 2013.
De Vries, F. T., Griffiths, R. I., Mark, B., Craig, H., Girlanda, M., Gweon, H. S., Hallin, S., Kaisermann, A., Keith, A. M., Kretzschmar, M., Lemanceau, P., Lumini, E., Mason, K. E., Oliver, A., Ostle, N., Prosser, J. I., Thion, C., Thomson, B., and Bardgett, R. D.: Soil bacterial networks are less stable under drought than fungal networks, Nat. Commun., 9, 3033, https://doi.org/10.1038/s41467-018-05516-7, 2018.
Edgar, R. C.: UPARSE: Highly accurate OTU sequences from microbial amplicon reads, Nat. Methods, 10, 996–998, 2013.
Egidi, E., Delgado-Baquerizo, M., Plett, J. M., Wang, J., Eldridge, D. J., Bardgett, R. D., Maestre, F. T., and Singh, B. K.: A few Ascomycota taxa dominate soil fungal communities worldwide, Nat. Commun., 10, 2369, https://doi.org/10.1038/s41467-019-10373-z, 2019.
Francisco-Ortega, J., Wang, Z. S., Wang, F. G., Xing, F. W., Liu, H., Xu, H., Xu, W. X., Luo, Y. B., Song, X. Q., Gale, S., Boufford, D. E., Maunder, M., and An, S. Q.: Seed plant endemism on Hainan Island: a framework for conservation actions, Bot. Rev., 76, 346–376, 2010.
Fuhrman, J. A.: Microbial community structure and its functional implications, Nature, 459, 193–199, 2009.
Geml, J., Timling, I., Robinson, C. H., Lennon, N., Nusbaum, H. C., Brochmann, C., Noordeloos, M. E., and Taylor, D. L.: An arctic community of symbiotic fungi assembled by dispersers: phylogenetic diversity of ectomycorrhizal basidiomycetes in Svalbard based on soil and sporocarp DNA, J. Biogeogr., 39, 74–88, 2012.
Gibson, L., Lee, T. M., Koh, L. P., Brook, B. W., Gardner, T. A., Barlow, J., Peres, C. A., Bradshaw, C. J. A., Laurance, W. F., Lovejoy, T. E., and Sodhi, N. S.: Primary forests are irreplaceable for sustaining tropical biodiversity, Nature, 478, 378–381, 2011.
Good, I. L.: The population frequencies of species and the estimation of population parameters, Biometrika, 40, 237–264, 1953.
Jiao, S., Liu, Z. S., Lin, Y. B., Yang, J., Chen, W. M., and Wei, G. H.: Bacterial communities in oil contaminated soils: biogeography and co-occurrence patterns, Soil Biol. Biochem., 98, 64–73, 2016.
Karimi, B., Maron, P. A., Chemidlin-Prevost Boure, N., Bernard, N., Gilbert, D., and Ranjard, L.: Microbial diversity and ecological networks as indicators of environmental quality, Environ. Chem. Lett., 15, 265–281, 2017.
Karimi, B., Dequiedt, S., Terrat, S., Jolivet, C., Arrouays, D., Wincker, P., Cruaud, C., Bispo, A., Prévost-Bouré, N. C., and Ranjard, L.: Biogeography of soil bacterial networks along a gradient of cropping intensity, Sci. Rep.-UK, 9, 3812, https://doi.org/10.1038/s41598-019-40422-y, 2019.
Kerfahi, D., Tripathi, B. M., Dong, K., Go, R., and Adams, J. M.: Rainforest conversion to rubber plantation may not result in lower soil diversity of bacteria fungi and nematodes, Microb. Ecol., 72, 359–371, 2016.
Kuperman, R. G., Siciliano, S. D., Römbke, J., and Oorts, K.: Deriving site-specific soil clean-up values for metals and metalloids: rationale for including protection of soil microbial processes, Integr. Environ. Assess., 10, 388–400, 2014.
Lan, G. Y., Li, Y. W., Wu, Z. X., and Xie, G. S.: Soil bacterial diversity impacted by conversion of secondary forest to rubber or eucalyptus plantations – a case study of Hainan Island, south China, Forest Sci., 63, 87–93, 2017a.
Lan, G. Y., Li, Y. W., Jatoi, M. T., Tan, Z. H., Wu, Z. X., and Xie, G. S.: Change in soil microbial community compositions and diversity following the conversion of tropical forest to rubber plantations in Xishuangbanan southwest China, Trop. Conserv. Sci., 10, 1–14, 2017b.
Lan, G. Y., Li, Y., Wu, Z. X., and Xie, G. S.: Impact of tropical forest conversion on soil bacterial diversity in tropical region of China, Eur. J. Soil Biol., 83, 91–97, 2017c.
Lan, G. Y., Li, Y. W., Lesueur, D., Wu, Z. X., and Xie, G. S.: Seasonal changes impact soil bacterial communities in a rubber plantation on Hainan Island China, Sci. Total Environ., 626, 826–834, 2018.
Lan, G. Y., Wu, Z. X., Sun, R., Yang, C., Chen, B. Q., and Zhang, X. C.: Tropical rainforest conversion into rubber plantations results in changes in soil fungal composition, but underling mechanisms of community assembly remain unchanged, Geoderma, 375, 114505, https://doi.org/10.1016/j.geoderma.2020.114505, 2020a.
Lan, G. Y., Wu, Z. X., Li, Y. W., and Chen, B. Q.: The drivers of soil bacterial communities in rubber plantation at local and geographic scales, Arch. Agron. Soil Sci., 66, 358–369, 2020b.
Lan, G. Y., Wu, Z. X., Sun, R., Yang, C., Chen, B. Q., and Zhang, X. C.: Forest conversion changed the structure and functional process of tropical forest soil microbiome, Land. Degrad. Dev., 32, 613–627, 2021.
Lopez, S., Rousset, F., Shaw, F. H., Ruth, G., Shaw, R. G., and Ophélie, R.: Joint effects of inbreeding and local adaptation on the evolution of genetic load after fragmentation, Conserv. Biol., 23, 1618–1627, 2009.
Louca, S., Polz, M. F., Mazel, F., Albright, M. B. N., Huber, J. A., O'Connor, M. I., Ackermann, M., Hahn, A. S., Srivastava, D. S., Crowe, S. A., Doebeli, M., and Parfrey, L. W.: Function and functional redundancy in microbial systems, Nat. Ecol. Evol., 2, 936–943, 2018.
Ma, H., Zou, W., Yang, J., Hogan, J. A., and Chen, J.: Dominant tree species shape soil microbial community via regulating assembly processes in planted subtropical forests, Forests, 10, 978, https://doi.org/10.3390/f10110978, 2019.
Mills, L. S., Soulé, M. E., and Doak, D. F.: The keystone-species concept in ecology and conservation, BioScience, 43, 219–224, 1993.
Morriën, E., Hannula, S. E., Snoek, L. B., Helmsing, N. R., Zweers, H., de Hollander, M., Soto, R. L., Bouffaud, M.-L., Buee, M., Dimmers, W., Duyts, H., Geisen, S., Girlanda, M., Griffiths, R. I., Jogensen, H.-B., Jensen, J., Plassart, P., Redecker, D., Schmelz, R. M., Schmidt, O., Thomson, B. C., Tisserant, E., Uroz, S., Winding, A., Bailey, M. J., Bonkowski, M., Faber, J. H., Martin, F., Lemanceau, P., de Boer, W., van Veen, J. A., and van der Putten, W. H.: Soil networks become more connected and take up more carbon as nature restoration progresses, Nat. Commun., 8, 14349, https://doi.org/10.1038/ncomms14349, 2017.
Mouquet, N., Gravel, D., Massol, F., and Calcagno, V.: Extending the concept of keystone species to communities and ecosystems, Ecol. Lett., 16, 1–8, 2013.
Neutel, A. M., Heesterbeek, J. A. P., and de Ruiter, P. C.: Stability in real food webs: weak links in long loops, Science, 296, 1120–1123, 2002.
Olesen, J. M., Bascompte, J., Dupont, Y. L., and Jordano, P.: The modularity of pollination networks, P. Natl. Acad. Sci. USA, 104, 19891–19896, 2007.
Pagliai, M., Vignozzi, N., and Pellegrini, S.: Soil structure and the effect of management practices, Soil Till. Res., 79, 131–143, 2004.
Philippot, L., Spor, A., Hénault, C., Bru, D., Bizouard, F., Jones, C. M., Sarr, A., and Maron, P. A.: Loss in microbial diversity affects nitrogen cycling in soil Laurent, ISME J., 7, 1609–1619, 2013.
Power, M., Tilman, D., Estes, J., Menge, B., Bond, W., Mills, S., Daily, G., Castilla, J., Lubchenco, J., Paine, R., Power, M., Tilman, D., Estes, J., Menge, B., Bond, W., Mills, L., Daily, G., Castilla, J., Lubchenco, J., and Paine, R.: Challenges in the quest for keystones, Bioscience, 46, 609–620, 1996.
Qin, J., Li, R., Raes, J., Arumugam, M., Burgdorf, K. S., Manichanh, C., Nielsen, T., Pons, N., Levenez, F., and Yamada, T.: A human gut microbial gene catalogue established by metagenomic sequencing, Nature, 464, 59–65, 2010.
Roger, G. and Amaral, L. A. N.: Functional cartography of complex metabolic networks, Nature, 433, 895, https://doi.org/10.1038/nature03288, 2005.
Rooney, N., McCann, K., Gellner, G., and Moore, J. C.: Structural asymmetry and the stability of diverse food webs, Nature, 442, 265–269, 2006.
Röttjers, L. and Faust, K.: From hairballs to hypotheses–biological insights from microbial networks, FEMS Microbiol. Rev., 10, 1093, https://doi.org/10.1093/femsre/fuy030, 2018.
Sahner, J., Budi, S. W., Barus, H., Edy, N., Meyer, M., Corré, M. D., and Polle, A.: Degradation of root community traits as indicator for transformation of tropical lowland rain forests into oil palm and rubber plantations, PloS One, 10, e0138077, https://doi.org/10.1371/journal.pone.0138077, 2015.
Sampson, T. R., Debelius, J. W., Thron, T., Janssen, S., Shastri, G. G., Ilhan, Z. E., Challis, C., Schretter, C. E., Rocha, S., Gradinaru, V., Chesselet, M. F., Keshavarzian, A., Shannon, K. M., Krajmalnik-Brown, R., Wittung-Stafshede, P., Knight, R., and Mazmanian, S. K.: Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson's disease, Cell, 167, 1469–1480, 2016.
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., Lesniewski, R. A., Oakley, B. B., Parks, D. H., and Robinson, C. J.: Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities, Appl. Environ. Microb., 75, 7537–7541, 2009.
Schneider, D., Engelhaupt, M., Allen, K., Kurniawan, S., Krashevska, V., Heinemann, M., and Scheu, S.: Impact of lowland rainforest transformation on diversity and composition of soil prokaryotic communities in Sumatra Indonesia, Front. Microbiol., 6, 296, https://doi.org/10.3389/fmicb.2015.01339, 2015.
Song, H., Singh, D., Tomlinson, K. W., Yang, X. D., Ogwu, M. C., Slik, J. W. F., and Adams, J. M.: Tropical forest conversion to rubber plantation in southwest China results in lower fungal beta diversity and reduced network complexity, FEMS Microbiol. Ecol., 95, fiz092, https://doi.org/10.1093/femsec/fiz092, 2019.
Stouffer, D. B. and Bascompte, J.: Compartmentalization increases food-web persistence, P. Natl. Acad. Sci. USA, 108, 3648–3652, 2011.
Taniguchi, T., Kitajima, K., Douhan, G. W., Yamanaka, N., and Allen, M. F.: A pulse of summer precipitation after the dry season triggers changes in ectomycorrhizal formation, diversity, and community composition in a Mediterranean forest in California, USA, Mycorrhiza, 28, 665–677, 2018.
Tripathi, B. M., Kim, M., Singh, D., Lee-Cruz, L., Lai-Hoe, A., Ainuddin, A. N., and Adams, J. M.: Tropical soil bacterial communities in Malaysia: pH dominates in the equatorial tropics too, Microb. Ecol., 64, 474–484, 2012.
Wagg, C., Schlaeppi, K., Banerjee, S., Kuramae, E. E., and van der Heijden, M. G. A.: Fungal-bacterial diversity and microbiome complexity predict ecosystem functioning, Nat. Commun., 10, 4841, https://doi.org/10.1038/s41467-019-12798-y, 2019.
Walters, W., Hyde, E. R., Berg-Lyons, D., Ackermann, G., Humphrey, G., Parada, A., Gilbert, J. A., Jansson, J. K., Caporaso, J. G., Fuhrman, J. A., Apprill, A., and Knight, R.: Improved bacterial 16S rRNA gene (V4 and V4-5) and fungal internal transcribed spacer marker gene primers for microbial community surveys, mSystems, 1, e00009-15, https://doi.org/10.1128/mSystems.00009-15, 2016.
Yang, W., Zhang, D., Cai, X., Xia, L., Luo, Y. Q., Cheng, X., and An, S. Q.: Significant alterations in soil fungal communities along a chronosequence of spartina alterniflora invasion in a Chinese yellow sea coastal wetland, Sci. Total Environ., 693, 133548, https://doi.org/10.1016/j.scitotenv.2019.07.354, 2019.
Zaller, J. G., Heigl, F., Ruess, L., and Grabmaier, A.: Glyphosate herbicide affects belowground interactions between earthworms and symbiotic mycorrhizal fungi in a model ecosystem, Sci. Rep., 4, 5634, https://doi.org/10.1038/srep05634, 2014.





