the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Microbial communities and their predictive functional profiles in the arid soil of Saudi Arabia
Munawwar A. Khan
Saudi Arabia has the world's fifth-largest desert and is the biggest importer of food and agricultural products. Understanding soil microbial communities is key to improving the agricultural potential of the region. Therefore, soil microbial communities of the semiarid region of Abha, known for agriculture, and arid regions of Hafar Al Batin and Muzahmiya were studied using Illumina sequencing. The results show that the microbial communities of the Saudi desert were characterized by the presence of high numbers of Actinobacteria, Proteobacteria, and Firmicutes. In addition to Sahara desert signature phyla like Gemmatimonadetes, biogeochemically important microorganisms like primary producers, nitrogen fixers and ammonia oxidizers were also present. The composition of the microbial community varied greatly among the sites sampled. The highest diversity was found in the rhizospheric soil of Muzahmiya followed by Abha. Firmicutes, Proteobacteria and Actinobacteria were the three main phyla detected in all the samples. Soils from the agricultural region of Abha were significantly different from other samples in containing only 1 % Firmicutes and 3–6 times higher population of Actinobacteria and Bacteroidetes, respectively. The presence of photosynthetic bacteria, ammonia oxidizers, and nitrogen fixers along with bacteria capable of surviving on simple and unlikely carbon sources like dimethylformamide was indicative of their survival strategies under harsh environmental conditions in the arid soil. Functional inference using PICRUSt analysis shows an abundance of genes involved in photosynthesis and nitrogen fixation.
- Article
(1058 KB) - Full-text XML
-
Supplement
(1030 KB) - BibTeX
- EndNote
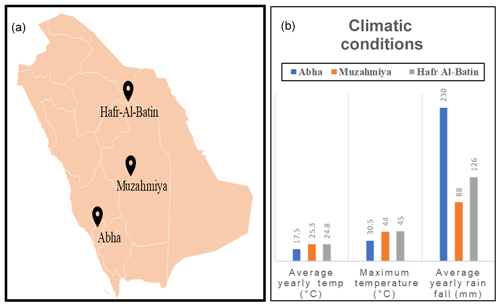
The Saudi Arabian Desert, also referred to as the Sahara Arabian Desert, is the fifth-largest desert of the world, bordering Yemen, the Persian Gulf, and Iraq (Holm, 1960). The desert is characterized by the presence of vast barren areas of sand referred to as empty quarters or Rub' al Khali and Wadi Al-Batin. Most of the area is barren with almost no vegetation, and the growing population is dependent on imported agricultural products for food (Fiaz et al., 2016). According to the government of the USA, Saudi Arabia imports USD 14.8 billion worth of agricultural products every year (https://www.export.gov/apex/article2?id=Saudi-Arabia-Market-Overview, last access: 19 October 2020). Although the climatic conditions are not favorable, the Saudi government has launched various programs to promote agriculture. In fact, 52.7 ×106 ha area, which is 25 % of the total country's area, is currently cultivable (Fiaz et al., 2016). Especially the Asir region, with Abha as its capital, is well known for agriculture and receives more rainfall than the rest of the country.
The microbial communities of arid regions are largely uncharacterized. To the best of our knowledge, no report from Saudi Arabia is available (Makhalanyane et al., 2015; Schulze-Makuch et al., 2018). The vast desert lacks vegetation and therefore is expected to be devoid of macromolecules and the microbial communities involved in the recycling of the nutrients. However, active microbial communities have been detected even in hyperarid deserts of the Atacama Desert, where rain is received only once per decade (Schulze-Makuch et al., 2018). Such studies are crucial in improving the agricultural potential in these extreme habitats and to design strategies for the modification of soil with microbial consortia for improving the agricultural potential of the arid soil (Fierer et al., 2012; Fierer, 2017). These microorganisms may alter soil fertility through sustaining the soil nutrient cycling, carbon sequestration, and by influencing other geochemical processes. This study, therefore, was aimed at comparing the soil microbial communities of Abha from the Asir region and arid regions of Muzahmiya and Hafar Al Batin. The knowledge of the microbial communities present in these regions may provide some insights into the role of microorganisms in various geochemical processes.
2.1 Soil sample collection
The desert soil samples were collected from three regions, namely Abha of Asir region (semiarid) and the arid regions of Muzahmiya (near Riyadh) and Hafar Al Batin. Samples were collected between 26 January and 18 February 2019. The weather of the city is generally mild throughout the year and is especially cooler during the “low-sun” season. The annual average temperature of Abha is only 17.5 ∘C and seldom rises above 35 ∘C. The city receives an annual rainfall of about 230 mm, most of which occurs between February and April and at an elevation of about 2270 m (7450 feet) above sea level. The soil type in Abha is sand and gravel, and the pH of the soil sample is slightly alkaline (i.e., 7.9). The region is known for agriculture, and the soil sample was collected from a plot with pristine soil. Muzahmiya is an arid region with an average annual temperature of 25.3 ∘C, and in summer it may cross 45 ∘C. The area receives annual precipitation of only 88 mm and is located at an elevation of 612 m above sea level. The soil type in Muzahmiya is reported to be Aridisol, sandy loam (Siham, 2007). Two samples were collected from Muzahmiya; the pH of both samples was alkaline (8.2). One sample was from the rhizospheric region of Haloxylon persicum, and another sample was collected from a distance of 1 m from the first sample. Hafar Al Batin is also an arid region with an average annual rainfall of only 126 mm and is located at an elevation of 358 m above sea level. The soil is Aridisol with an alkaline pH of 8.4. Figure 1 and Table 1 show the locations of the sampling sites and the climatic conditions at these sites. Three cores of 1.9 cm diameter from each sampling site were collected from a distance of 1 m from each other and were mixed to obtain a composite sample. Debris (2 cm) from the surface was removed at the time of sampling, and a soil core from a depth of 5 cm was then obtained. From Muzahmiya two samples – namely a rhizospheric soil and a non-rhizospheric soil sample – were collected. Following collection, samples were transported to the lab at room temperature and were homogenized via sieving (<2 mm). Portions of soils were stored at −20 ∘C for DNA extraction. Biogeochemical properties and other details of sampling sites are given in Table 1. The soil pH was determined in 0.01 M CaCl2 (2:1 solution to solid ratio) using a pH meter, while total aerobic counts in the soil were determined on one-fifth diluted nutrient agar plates using the dilution plating method.
2.2 DNA extraction and HiSeq analysis
Genomic DNA from the soil was prepared using the direct lysis method of Robe et al. (2003). For obtaining enough DNA, extraction was carried out in replicates, and the DNA was pooled and concentrated. The composition and diversity of bacterial communities in soil were determined by amplifying the V3–V4 regions of bacterial 16S ribosomal RNA (rRNA) genes. A set of 341F and 806R primers and the DNA extracted from the samples as a template were used for amplification. PCRs were carried out at 50 µL scale containing ∼40 ng of DNA template, 25 µL DreamTaq Green PCR Master Mix (2×), 20.5 µL H2O, 0.5 µL of 1 % bovine serum albumin, and 0.2 µM of each primer. The PCR was carried out by programming the thermal regime as initial denaturation at 95 ∘C for 5 min – followed by 35 cycles of denaturation at 95 ∘C for 30 s, annealing at 56 ∘C for 30 s, and extension at 72 ∘C for 30 s, and a final extension step at 72 ∘C for 7 min. Amplicon sequencing was conducted on an Illumina HiSeq 2500 platform. Further processing of the reads and quality filtering was conducted as described earlier (Yuan et al., 2018).
2.3 Data analysis
Raw data obtained from the sequencing were processed using QIIME (Quantitative Insights Into Microbial Ecology; Caporaso et al., 2010). Sequences were clustered into operational taxonomic units (OTUs) using UCLUST and an identity threshold of 97 %. Sequences were assigned to their phylogenetic groups using the QIIME pipeline and the Greengenes Database version 13.5 (Santamaria et al., 2012). Further processing of the sequences was carried out using Calypso (Zakrzewski et al., 2017). Rarefaction curves were calculated for the number of species present in each sample. Alpha diversity was determined using both taxonomic metrics (numbers of phylotypes). To test whether sample categories harbored significantly different microbial communities, we used an analysis of similarities (ANOSIM). To determine whether the relative abundances of individual taxa were significantly different between sample categories, pairwise t tests with P values were calculated. An abundance of various taxa in the samples as correlation charts were also calculated using Calypso, while abundance pie charts were calculated using Krona (Ondov et al., 2013). Predictive functional analysis of microbial communities using 16S rRNA gene sequences was carried out using PICRUSt (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States), and STAMP (statistical analysis of taxonomic and functional profiles) for functional inference using PICRUSt and STAMP software v 2.1.3 was used for statistical analyses and to detect differentially abundant OTUs between two sample groups (Parks et al., 2014).
The sequences have been submitted to the Sequence Read Archive (SRA) with the accession numbers SAMN12651127–12651133.
3.1 Properties of soil samples
The details of the collected soil samples are given in Table 1. The pH of all the soil samples was alkaline and ranged from 7.9 to 8.4. The soil type in Abha is sand and gravel, and the pH of the soil sample was slightly alkaline (i.e., 7.9). The total CFU count obtained on nutrient agar medium was CFU g−1 of soil. The soil type in Muzahmiya is reported to be sandy loam (Siham, 2007). The CFU count of the rhizospheric soil of Haloxylon persicum was 50 times higher ( CFU g−1 of soil) than the non-rhizospheric ( CFU g−1 of soil) soil at a distance of a few centimeters. Hafar Al Batin is also an arid region with an average annual rainfall of only 126 mm. The soil was alkaline with a pH of 8.4, and the aerobic bacterial count on nutrient agar was CFU g−1 of soil.
3.2 Microbial diversity of soil samples
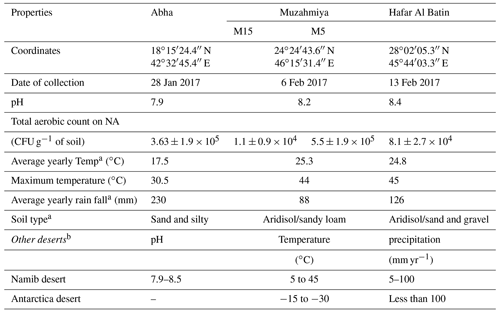
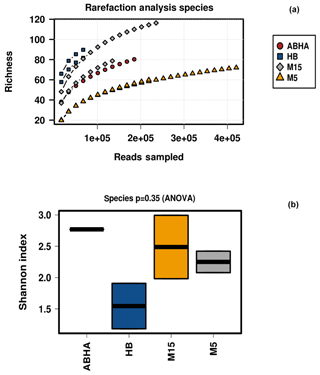
Reads per sample varied between 68 640 and 420 570. For analysis, a total of 12 790 data points were included for each sample. The alpha diversity in the samples was calculated using species as OTUs. Rarefaction curves and Shannon diversity index show the highest diversity is associated with the samples collected from the rhizospheric soil (M15) of Haloxylon persicum collected from Muzahmiya (Fig. 2). The microbial population was least diverse in the sample collected from the non-rhizospheric soil from the same region (M5). It can also be noted that the diversity of rhizospheric soil from Muzahmiya was comparable to the diversity observed in the samples collected from Abha, a region known for agriculture. Aerobic plate counts obtained from the two samples were also comparable. The species richness of Hafar Al Batin (HB) was also comparable to the samples collected from Abha and Muzahmiya, although the aerobic plate count was as low as that found in non-rhizospheric soil from Muzahmiya (M5). R values of 0.85 obtained in ANOSIM show that the studied microbial communities are significantly different from each other (Fig. 3a).
Figure 3Analysis of similarity (ANOSIM) within and between samples (a). The abundance of different phyla in desert soil samples studied and their comparison with the hot (Namib) and cold (Antarctica) desert samples are shown in (b). Data for Antarctic desert and Namib desert are taken from Armstrong et al. (2016) and Cary et al. (2010), respectively.
Between 69 % and 88 % of the total reads were assigned to Bacteria, and the majority of the reads can be assigned to the phyla Actinobacteria, Firmicutes and Proteobacteria, while remaining reads were assigned to unknown groups detailed in Supplement Fig. S1. Populations of Firmicutes in samples collected from Muzahmiya M15, M5, Hafar Al Batin and Abha were 50 %, 0.7 %, 13 % and 1 % of the total bacteria, respectively, while the population of Proteobacteria was 25 %, 90 %, 31 % and 53 % in soil samples from Muzahmiya M15, M5, Hafar Al Batin and Abha, respectively. The population of Actinobacteria was highest in soil from Abha (31 % of the total bacteria), while a comparable population of Actinobacteria in soils of Hafar Al Batin (10 %) and rhizospheric soil of Muzahmiya (9 %) was observed (Fig. 3b). Earlier reports show that the major soil bacteria found in desert soil belong to Actinobacteria, Bacteroidetes and Proteobacteria (Fierer et al., 2012; Andrew et al., 2012). Interestingly, a recent study of Saudi desert soil samples also shows the presence of Actinobacteria, Bacteroidetes and Proteobacteria as the major soil phyla (Eida et al., 2018). A soil sample from Abha shows almost the same pattern where the population of Proteobacteria, Actinobacteria and Bacteroidetes was 53 %, 31 % and 6 % of the total bacteria, respectively, while samples from Muzahmiya and Hafar Al Batin vary in not having the significant populations of Bacteroidetes and an increased population of Firmicutes (Fig. 3b). A high population of Bacteroidetes in samples other than Abha may be due to the unavailability of complex organic matter in these soils. Members of the Bacteroidetes are known to degrade various macromolecules in the soil. The high populations of Firmicutes and Actinobacteria in desert soil may be due to their ability to produce spores under high temperature and aridity. It is to be noted that temperatures do not exceed 35 ∘C in Abha, which has the lowest population of Firmicutes, which may not be the case with other deserts. A high population of Actinobacteria has been found in both the cold Antarctica desert as well as hot Namib desert (Aislabie et al., 2006; Armstrong et al., 2016). Interestingly, the highest population of Actinobacteria (34 % of the total bacteria) was observed in Abha, which is geographically close to the Namib desert. Acidobacteria were only found in the rhizospheric soil of Muzahmiya (2 % of the total), while Planctomycetes were found in the soils of Abha and Hafar Al Batin. Among other minor phyla, notably present in soil samples were the members of phylum Gemmatimonadetes, like Gemm 3 in some cases constituting as much as 4 % (Muzahmiya rhizospheric soil) of the total reads. These bacteria have been reported earlier also in the desert soil, and recently strains from the phylum with photosynthetic capability have been cultured (Meola et al., 2015; Zeng et al., 2014). The ammonia-oxidizing Archaea Candidatus Nitrososphaera gargensis was also found in most of the samples. Reads belonging to iii115 constituted up to 3 % of the total population at least in two samples (M15 and Hafar Al Batin); these sequences have also been reported from the soil in earlier studies (Marasco et al., 2018). The Antarctica desert soil survey shows that Actinobacteria were present prominently along with Bacillus spp., Flavobacterium spp. and Acinetobacter spp. Deinococcus–Thermus and Gemmatimonadetes clades, which have low or no representation in other surface soils. They are relatively common in dry valley clone libraries. Members of 13 phyla have been found in the soil of Antarctica desert including Actinobacteria, Gemmatimonas, Proteobacteria, Bacteroidetes, Deinococcus and Thermus, Planctomycetes, Chloroflexi, Verrucomicrobia, Acidobacteria, Cyanobacteria, TM7, and OP11. The most dominant were Acidobacteria, Actinobacteria and Bacteroidetes (Cary et al., 2010). In the case of the hot Namib desert, 19 different phyla were observed, as shown in Fig. 3b. The most abundant phyla were Bacteriodetes, Proteobacteria, and Actinobacteria (Armstrong et al., 2016).
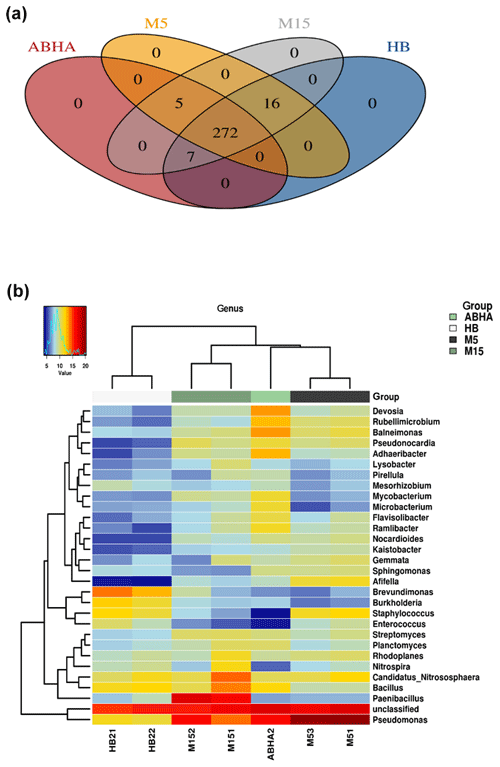
Figure 4Venn diagram showing the number of genera present in different desert soil samples (a). Heatmap showing the 20 most dominant genera present in the desert soil samples. M5 and M15: samples collected from Muzahmiya, HB: sample collected from Hafar Al Batin.
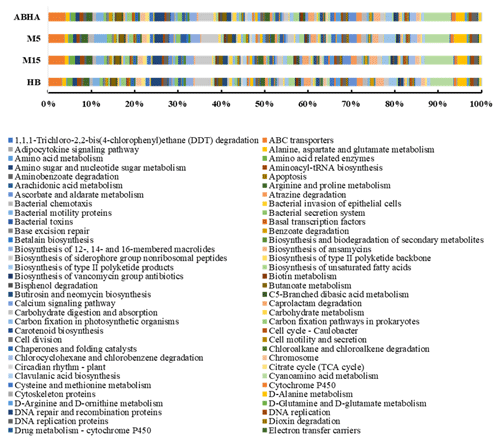
Figure 5Predicted functional gene abundance in four different samples obtained through PICRUSt analysis. M5 and M15: samples collected from Muzahmiya, HB: sample collected from Hafar Al Batin.
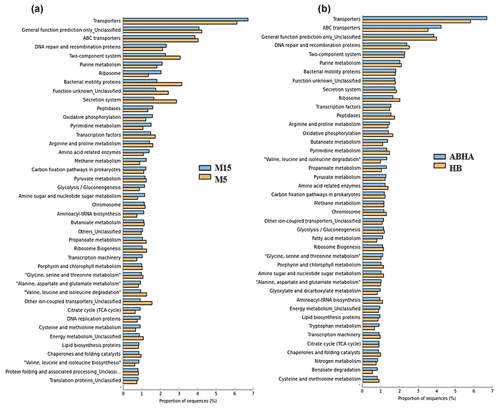
Figure 6Statistical comparison (Welch's t test) of the predicted function gene abundance between M5 and M15 samples obtained from Muzahmiya (a) and between ABHA and Hafar Al Batin (b). P-value correction; P<0.05.
The relative abundance of the major genus found in the soil samples is shown as a heatmap in Fig. 4b. A detailed microbial community composition generated by the Calypso program is shown in Figs. S1–S4. The Venn diagram (Fig. 4a) shows that the core genera found in all the samples were 272, while Abha and rhizospheric soil sample from Muzahmiya (M15) shared the maximum number of genera (300), while M15 and HB also shared 295 genera and Abha and HB shared 279 genera. Some of the most abundant genera include Pseudomonas, Paenibacillus, Bacillus, Candidatus Nitrososphaera, Devosia, Adhaeribacter and others. The bacterial genera found in desert soil are expected to withstand extreme climatic conditions and to perform some vital functions. For example, genera like Bacillus and Paenibacillus or Actinobacteria like Nocardioides, and Streptomyces are spore-forming bacteria and hence can survive extreme heat and arid conditions. Many of these genera have been isolated from the desert already. Ramlibacter, one of the genera found in our samples, forms cyst, and its genome analysis shows adaptation to arid conditions (De Luca et al., 2011). Ramlibacter was originally isolated from the meteorite fragments buried in the sands of a desert (De Luca et al., 2011). Modestobacter, another dominant bacteria found in our samples, was isolated from the Atacama Desert of Chile, South America (Busarakam et al., 2016). Genera like Pseudomonas and Adhaeribacter may produce extracellular polysaccharide to survive under arid conditions and to form strong biofilms. They may also contribute to water retention in soil promoting the formation of soil crust.
It should be noted that some bacteria may play important roles besides tolerating the extreme conditions. Candidatus Nitrososphaera and Planctomyces found in all the samples in high numbers are known to oxidize ammonia (Stieglmeier et al., 2014). Notably, it was observed that the desert soil catalyzes ammonia formation (Schrauzer, 1978). Nitrite-oxidizing bacteria Nitrospira were also found in all the samples, but the reads were especially high in rhizospheric soil from Muzahmiya. The phototrophic bacterium Rhodoplanes present in all the samples may be one of the bacteria involved in carbon fixation in the nutrient-deficient arid soil. Members of Devosia are known to nodulate Neptunia for nitrogen fixation and, therefore, may serve as a nitrogen source in otherwise nitrogen-deficient arid soils. Members of the genera Mesorhizobium, Bradyrhizobium and Sinorhizobium were also found in all the samples, while the members of the genus Rhizobium were detected only in rhizospheric soil from Muzahmiya. Another genus found in significant numbers in all the soil samples and whose population was especially high in Abha was Balneimonas, which is a member of family Bradyrhizobiaceae, known to produce extracellular material that plays an important role in the formation of soil crust (Matthews et al., 2019).
Abha soil sample was also distinct in having high populations of bacteria with the ability to survive on the simple source of nutrients and extreme conditions or the ability to perform an important geochemical or agricultural function. The population of genera Adhaeribacter, Modestobacter, Ramlibacter, radiation-resistant Geodermatophilus, Pseudonocardia and Flavobacterium was high in samples from Abha. The population of Paracoccus and Phenylobacterium capable of growing optimally on artificial compounds like dimethylformamide, chloridazon, antipyrin and pyramidon was also significantly higher (Eberspächer and Lingens, 2006). N2-fixing Azospirillum, Agrobacterium and phototrophic Rhodobacter were also present in high numbers in soil from Abha, while the rhizospheric soil of Muzahmiya also shows a similar pattern containing a high population of Candidatus Nitrososphaera, Ramlibacter, Bradyrhizobium and phototrophic Rhodoplanes. But the sample was different in having significantly high populations of bacteria like Paenibacillus, Alicyclobacillus and Sporosarcina. Soil samples collected from Hafar Al Batin have a completely different community with high populations of Pseudomonas, Propionibacterium, Brevundimonas, Staphylococcus and Burkholderia. The microbial community in Hafar Al Batin soil is completely different probably due to the completely different environmental conditions in Hafar Al Batin. This region is well known for its extreme arid conditions in Saudi Arabia. The functional inference using PICRUSt analysis shows similar results (Figs. 5 and 6). Some of the most abundant genes belong to transporters, peptidases, housekeeping genes and general function. Genes involved in prokaryotic photosynthesis and chlorophyll metabolism constitute more than 2 % of the total genes (Figs. 5 and 6). Furthermore, genes involved in the metabolism of simple substrates like methane, butanoate and benzoate were also predicted to have a high proportion. This indicates the survival strategy of the microbial community under nutrient-deficient harsh environmental conditions. Interestingly, it was found that the proportion of genes for prokaryotic photosynthesis was lowest (1∕4th) in samples from Abha compared to other samples. Probably comparatively higher soil fertility and the semiarid nature of soil do not require a high population of photosynthetic bacteria for maintaining and providing carbon to other soil organisms. Similarly, the proportion of genes involved in methane and nitrogen metabolism and peptidoglycan biosynthesis were lowest (rd) in Abha samples. Indicating that the population of Gram-positive bacteria in all the samples other than the sample from Abha is high. Production of methane is a characteristic of arid soils, and the presence of these genes in high proportions in all the samples other than Abha further confirms the fact observed in previous studies.
Understanding the composition of the desert microbial communities may help us in understanding the role of different microorganisms in extreme environments. The analysis shows that the microbial communities of the Saudi desert were characterized by the presence of high numbers of Actinobacteria, Proteobacteria and Firmicutes. These microbial communities, besides showing Saharan desert signature phyla like Gemmatimonas, also show biogeochemically important microorganisms exemplified by primary producers like Rhodoplanes and Cyanobacteria, nitrogen-fixing members of the genus Rhizobium and Bradyrhizobium, and ammonia oxidizer Candidatus Nitrososphaera. Communities were also characterized by the presence of microbes capable of growing on simple and unlikely carbon sources such as methane, butanoate and dimethylformamide, indicating the survival strategies adopted by microbial communities under nutrient-deficient conditions.
The nucleotide sequence data have been submitted to GenBank with the accession numbers SAMN12651127–12651133 (https://www.ncbi.nlm.nih.gov/, last access: 20 October 2020).
Figures S1–S4 show the diversity of microbial community as Krona pie charts generated using Calypso for M15, M5, Abha and Hafar Al Batin, respectively. The supplement related to this article is available online at: https://doi.org/10.5194/soil-6-513-2020-supplement.
MAK performed the data analysis and prepared the manuscript. STK collected the samples, prepared genomic DNA and carried out sequencing.
The authors declare that they have no conflict of interest.
We would like to thank the Zayed University Office of Research for financial support.
This research has been supported by the Zayed University Microbial Diversity and Eco-genomics research cluster project (grant no. R16092).
This paper was edited by Fuensanta García-Orenes and reviewed by three anonymous referees.
Aislabie, J. M., Chhour, K.-L., Saul, D. J., Miyauchi, S., Ayton, J., Paetzold, R. F., and Balks, M. R.: Dominant bacteria in soils of Marble Point and Wright Valley, Victoria Land, Antarctica, Soil Biol. Biochem., 38, 3041–3056, https://doi.org/10.1016/j.soilbio.2006.02.018, 2006.
Al-Zahrani, M., Al-Areeq, A., and Sharif, H. O.: Estimating urban flooding potential near the outlet of an arid catchment in Saudi Arabia, Geomatics, Natural Hazards and Risk, 8, 672–688, 2017.
Andrew, D. R., Fitak, R. R., Munguia-Vega, A., Racolta, A., Martinson, V. G., and Dontsova, K.: Abiotic Factors Shape Microbial Diversity in Sonoran Desert Soils, Appl. Environ. Microbiol., 78, 7527–7537, https://doi.org/10.1128/AEM.01459-12, 2012.
Armstrong, A., Valverde, A., Ramond, J.-B., Makhalanyane, T., Jansson, J., Hopkins, D., Aspray, T., Seely, M., Tuffin, M., and Cowan, D.: Temporal dynamics of hot desert microbial communities reveal structural and functional responses to water input, Sci. Rep., 6, 34434, https://doi.org/10.1038/srep34434, 2016.
Busarakam, K., Bull, A. T., Trujillo, M. E., Riesco, R., Sangal, V., van Wezel, G. P., and Goodfellow, M.: Modestobacter caceresii sp. nov., novel actinobacteria with an insight into their adaptive mechanisms for survival in extreme hyper-arid Atacama Desert soils, Syst. Appl. Microbiol., 39, 243–251, https://doi.org/10.1016/j.syapm.2016.03.007, 2016.
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., Fierer, N., Pena, A. G., Goodrich, J. K., Gordon, J. I., Huttley, G. A., Kelley, S. T., Knights, D., Koenig, J. E., Ley, R. E., Lozupone, C. A., McDonald, D., Muegge, B. D., Pirrung, M., Reeder, J., Sevinsky, J. R., Turnbaugh, P. J., Walters, W. A., Widmann, J., Yatsunenko, T., Zaneveld, J., and Knight, R.: QIIME allows analysis of high-throughput community sequencing data, Nat. Method., 7, 335–336, https://doi.org/10.1038/nmeth.f.303, 2010.
Cary, S. C., McDonald, I. R., Barrett, J. E., and Cowan, D. A.: On the rocks: the microbiology of Antarctic Dry Valley soils, Nat. Rev. Microbiol., 8, 129–138, https://doi.org/10.1038/nrmicro2281, 2010.
De Luca, G., Barakat, M., Ortet, P., Fochesato, S., Jourlin-Castelli, C., Ansaldi, M., Py, B., Fichant, G., Coutinho, P. M., Voulhoux, R., Bastien, O., Marechal, E., Henrissat, B., Quentin, Y., Noirot, P., Filloux, A., Mejean, V., DuBow, M. S., Barras, F., Barbe, V., Weissenbach, J., Mihalcescu, I., Vermeglio, A., Achouak, W., and Heulin, T.: The Cyst-Dividing Bacterium Ramlibacter tataouinensis TTB310 Genome Reveals a Well-Stocked Toolbox for Adaptation to a Desert Environment, PLoS ONE, 6, e23784, https://doi.org/10.1371/journal.pone.0023784.g001, 2011.
Eberspächer, J. and Lingens, F.: The Genus Phenylobacterium, in: The Prokaryotes: Vol. 5, Proteobacteria: Alpha and Beta Subclasses, edited by: Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.-H., and Stackebrandt, E., Springer New York, New York, NY, 250–256, 2006.
Eida, A. A., Ziegler, M., Lafi, F. F., Michell, C. T., Voolstra, C. R., Hirt, H., and Saad, M. M.: Desert plant bacteria reveal host influence and beneficial plant growth properties, PLoS One, 13, e0208223, https://doi.org/10.1371/journal.pone.0208223, 2018.
Fiaz, S., Noor, M. A., and Aldosari, F.: Achieving food security in the Kingdom of Saudi Arabia through innovation: Potential role of agricultural extension, J. Saudi Soc. Agr. Sci., 17, 365–375, https://doi.org/10.1016/j.jssas.2016.09.001, 2016.
Fierer, N.: Embracing the unknown: disentangling the complexities of the soil microbiome, Nat. Rev. Microbiol., 15, 579–590, https://doi.org/10.1038/nrmicro.2017.87, 2017.
Fierer, N., Leff, J. W., Adams, B. J., Nielsen, U. N., Bates, S. T., Lauber, C. L., Owens, S., Gilbert, J. A., Wall, D. H., and Caporaso, J. G.: Cross-biome metagenomic analyses of soil microbial communities and their functional attributes, P. Natl. Acad. Sci. USA, 109, 21390–21395, https://doi.org/10.1073/pnas.1215210110, 2012.
Holm, D. A.: Desert Geomorphology in the Arabian Peninsula, Science, 132, 1369–1379, https://doi.org/10.1126/science.132.3437.1369, 1960.
Makhalanyane, T. P., Valverde, A., Gunnigle, E., Frossard, A., Ramond, J. B., and Cowan, D. A.: Microbial ecology of hot desert edaphic systems, FEMS Microbiol. Rev., 39, 203–221, https://doi.org/10.1093/femsre/fuu011, 2015.
Marasco, R., Rolli, E., Fusi, M., Michoud, G., and Daffonchio, D.: Grapevine rootstocks shape underground bacterial microbiome and networking but not potential functionality, Microbiome, 6, 3, https://doi.org/10.1186/s40168-017-0391-2, 2018.
Matthews, A., Pierce, S., Hipperson, H., and Raymond, B.: Rhizobacterial Community Assembly Patterns Vary Between Crop Species, Front Microbiol., 581, 10, https://doi.org/10.3389/fmicb.2019.00581, 2019.
Meola, M., Lazzaro, A., and Zeyer, J.: Bacterial Composition and Survival on Sahara Dust Particles Transported to the European Alps, Front. Microbiol., 6, 1454–1454, https://doi.org/10.3389/fmicb.2015.01454, 2015.
Ondov, B. D., Bergman, N. H., and Phillippy, A. M.: Krona: Interactive Metagenomic Visualization in a Web Browser, in: Encyclopedia of Metagenomics, edited by: Nelson, K. E., Springer New York, New York, NY, 1–8, 2013.
Parks, D. H., Tyson, G. W., Hugenholtz, P., and Beiko, R. G.: STAMP: statistical analysis of taxonomic and functional profiles, Bioinformatics, 30, 3123–3124, 10.1093/bioinformatics/btu494, 2014.
Robe, P., Nalin, R., Capellano, C., Vogel, T. M., and Simonet, P.: Extraction of DNA from soil, Europ. J. Soil Biol., 39, 183–190, https://doi.org/10.1016/S1164-5563(03)00033-5, 2003.
Santamaria, M., Fosso, B., Consiglio, A., De Caro, G., Grillo, G., Licciulli, F., Liuni, S., Marzano, M., Alonso-Alemany, D., Valiente, G., and Pesole, G.: Reference databases for taxonomic assignment in metagenomics, Brief. Bioinform., 13, 682–695, https://doi.org/10.1093/bib/bbs036, 2012.
Schrauzer, D. G. N.: Desert sands catalyze ammonia formation, Chem. Eng. News Arch., 56, 7 pp., https://doi.org/10.1021/cen-v056n046.p007a, 1978.
Schulze-Makuch, D., Wagner, D., Kounaves, S. P., Mangelsdorf, K., Devine, K. G., de Vera, J.-P., Schmitt-Kopplin, P., Grossart, H.-P., Parro, V., Kaupenjohann, M., Galy, A., Schneider, B., Airo, A., Frösler, J., Davila, A. F., Arens, F. L., Cáceres, L., Cornejo, F. S., Carrizo, D., Dartnell, L., DiRuggiero, J., Flury, M., Ganzert, L., Gessner, M. O., Grathwohl, P., Guan, L., Heinz, J., Hess, M., Keppler, F., Maus, D., McKay, C. P., Meckenstock, R. U., Montgomery, W., Oberlin, E. A., Probst, A. J., Sáenz, J. S., Sattler, T., Schirmack, J., Sephton, M. A., Schloter, M., Uhl, J., Valenzuela, B., Vestergaard, G., Wörmer, L., and Zamorano, P.: Transitory microbial habitat in the hyperarid Atacama Desert, P. Natl. Acad. Sci. USA, 115, 2670–2675, https://doi.org/10.1073/pnas.1714341115, 2018.
Siham, A.-K.: Soil Analysis of Contaminated Soil from Riyadh City, Saudi Arabia and Influence of Aluminium and Cobalt Ions on the Growth of Fungi Isolated, J. Biol. Sci., 7, https://doi.org/10.3923/jbs.2007.549.553, 2007.
Stieglmeier, M., Klingl, A., Alves, R. J., Rittmann, S. K., Melcher, M., Leisch, N., and Schleper, C.: Nitrososphaera viennensis gen. nov., sp. nov., an aerobic and mesophilic, ammonia-oxidizing archaeon from soil and a member of the archaeal phylum Thaumarchaeota, Int. J. Syst. Evol. Micr., 64, 2738–2752, https://doi.org/10.1099/ijs.0.063172-0, 2014.
Yuan, H.-Y., Liu, P.-P., Wang, N., Li, X.-M., Zhu, Y.-G., Khan, S. T., Alkhedhairy, A. A., and Sun, G.-X.: The influence of soil properties and geographical distance on the bacterial community compositions of paddy soils enriched on SMFC anodes, J. Soil. Sediment., 18, 517–525, https://doi.org/10.1007/s11368-017-1769-2, 2018.
Zakrzewski, M., Proietti, C., Ellis, J. J., Hasan, S., Brion, M. J., Berger, B., and Krause, L.: Calypso: a user-friendly web-server for mining and visualizing microbiome-environment interactions, Bioinformatics, 33, 782–783, https://doi.org/10.1093/bioinformatics/btw725, 2017.
Zeng, Y., Feng, F., Medova, H., Dean, J., and Koblizek, M.: Functional type 2 photosynthetic reaction centers found in the rare bacterial phylum Gemmatimonadetes, P. Natl. Acad. Sci. USA, 111, 7795–7800, https://doi.org/10.1073/pnas.1400295111, 2014.