the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Acute glyphosate exposure does not condition the response of microbial communities to a dry–rewetting disturbance in a soil with a long history of glyphosate-based herbicides
Elena Gomez
María Celina Zabaloy
Dry–rewetting perturbations are natural disturbances in the edaphic environment and particularly in dryland cultivation areas. The interaction of this disturbance with glyphosate-based herbicides (GBHs) deserves special attention in the soil environment due to the intensification of agricultural practices and the acceleration of climate change with an intensified water cycle. The objective of this study was to assess the response of microbial communities in a soil with a long history of GBHs to a secondary imposed perturbation (a single dry–rewetting event). A factorial microcosm study was conducted to evaluate the potential conditioning effect of an acute glyphosate exposure on the response to a following dry–rewetting event. A respiratory quotient (RQ) based on an ecologically relevant substrate (p-coumaric acid) and basal respiration was used as a physiological indicator. Similarly, DNA-based analyses were considered, including quantitative PCR (qPCR) of functional sensitive microbial groups linked to cycles of carbon (Actinobacteria) and nitrogen (ammonia-oxidizing microorganisms), qPCR of total bacteria and denaturing gradient gel electrophoresis (DGGE) of ammonia-oxidizing bacteria (AOB). Significant effects of herbicide and of dry–rewetting perturbations were observed in the RQ and in the copy number of the amoA gene of AOB, respectively. However, no significant interaction was observed between them when analyzing the physiological indicator and the copy number of the evaluated genes. PCR–DGGE results were not conclusive regarding a potential effect of dry–rewetting × herbicide interaction on AOB community structure, suggesting further analysis by deep sequencing of the amoA gene. The results of this study indicate that the perturbation of an acute glyphosate exposure in a soil with a long history of this herbicide does not have a conditioning effect on the response to a subsequent dry–rewetting disturbance according to a physiological indicator or the quantified bacterial/archaeal genes. This is particularly relevant for the sustainability of soils in rainfed agriculture, where frequent exposure to GBHs along with intensification of hydrological cycles are expected to occur. Further studies considering multiple dry–rewetting disturbances and in different soil types should be conducted to simulate those conditions and to validate our results.
- Article
(442 KB) - Full-text XML
- BibTeX
- EndNote
Soil microbial communities play a central role in several processes that contribute to a wide range of important ecosystem services (Tilman et al., 2002; EFSA Panel on Plant Protection Products and their Residues, 2016). Different factors with potential disruption effects on microbial communities and processes can reduce the functional sustainability of soils (Tilman et al., 2002). Among them, anthropic disturbances (e.g. pesticides) or natural disturbances like dry–rewetting events are common perturbations of the soil environment, particularly in the context of global climate models which predict an intensification of the hydrological cycles with more extended periods of droughts and more intense rainfalls (Huntington, 2006).
The effects of dry–rewetting cycles in the edaphic environment and on microbial communities have been considered in several studies (Hastings et al., 2000; Gleeson et al., 2008; Bustamante et al., 2012). Desiccation can affect microbial communities through nutritional limitation, osmotic stress and competition for available nutrients (Griffiths et al., 2003). Similarly, a rapid rewetting can trigger an osmotic shock-inducing lysis, release of intracellular solutes and an increase in C and N mineralization (Fierer et al., 2003). However, the interaction of these disturbances with the perturbation imposed by glyphosate-based herbicides (GBHs) has not been assessed before, even when the simultaneous exposure to both factors represents a common scenario in dryland cultivation areas such as in the semiarid Pampas of Argentina. These disturbance events could increase their frequency due to the intensification of agricultural practices based on glyphosate-resistant (GR) crops (Cerdeira and Duke, 2006) and repeated dry–rewetting cycles under an accelerating climate change (Huntington, 2006; Evans and Wallenstein, 2011).
In a previous study, we reported no detection of a pollution-induced increase in microbial community tolerance (PICT) to glyphosate in a soil with a long history of GBHs (Allegrini et al., 2015). Considering the aforementioned, we conducted a follow-up study to assess the response of microbial communities of a soil chronically exposed to GBHs to a secondary imposed perturbation (a single dry–rewetting event). The response of microbial communities to the perturbations imposed by glyphosate exposure and dry–rewetting was assessed through a physiological indicator, calculated as the ratio of basal respiration to substrate-induced respiration (SIR) with p-coumaric acid as the amended substrate. This respiratory quotient (RQ) has proven to be sensitive to repeated glyphosate applications (Allegrini et al., 2017). Similarly, DNA-based analyses were conducted to quantify the abundance of genes from different microbial groups which could be affected by the imposed perturbations. We focused on microorganisms with well-known sensitivity to GBHs and other pesticides like ammonia-oxidizing bacteria and archaea (AOB, AOA) (Zhang et al., 2018) and Actinobacteria (Barriuso et al., 2010). Ammonia-oxidizing prokaryotes and actinomycetes are involved in ecologically relevant processes in soil (N cycling and organic matter turnover, respectively) and have been classified as microorganisms with a high degree of sensitivity with respect to losses of organisms or functions (Anderson, 2003). We hypothesize that, if no increase in community tolerance was observed after long exposure to GBHs in the field, an acute exposure would not significantly modify the structure and physiology of the microbial community so as to condition the sensitivity to a subsequent dry–rewetting disturbance.
2.1 Soil sampling and microcosm set-up
Sampling was conducted in the same agricultural plot (ZAVH; 33∘02′23′′ S, 60∘53′05′′ W) with a long history (more than 20 years) of exposure to GBHs that was described in a previous study (Allegrini et al., 2015). The soil is a Vertic Argiudoll with clay silt loam to clay silt texture. Fifteen subsamples were taken at a 0–10 cm depth, sieved (<5.6 mm) and pooled to obtain a composite sample. Soil was stored at 4 ∘C and used within 6 d for the microcosm study.
Twelve microcosms (equivalent to 40 g of oven dry soil) were prepared in 100 mL sterile screw-cap polypropylene flasks, loosely capped to reduce water evaporation whilst leaving enough space for free passage of air. All flasks (60 % WHC) were pre-incubated in the dark at 25 ∘C (Ingelab I.501PF Incubator) for 1 week. Then, microcosms were randomly assigned to the following treatments, in a 2×2 factorial design with three replicates per treatment: “herbicide” (two levels: with GBH “CG” and control with distilled sterile water “SG”) and “dry–rewetting” (two levels: with desiccation “CD” and untreated control “SD”). First, microcosms received either the CG or SG treatment (day 0). The herbicide (Roundup Full II, Monsanto™, N–(phosphonomethyl)glycine potassium salt, 66.2 % w∕v, additives not specified) was applied in a final volume of 0.2 mL (with distilled water) at a rate of 49 µg active ingredient g−1 soil similarly to other studies with silt loam soils (Haney et al., 2000; Ratcliff et al., 2006). This dose mimics the concentration of glyphosate found in soil after a 1 × application rate in the field (0.84 kg ha−1) considering a 2 mm soil interaction penetration due to the high absorptivity and low leachability of glyphosate (Haney et al., 2000). Microcosms were initially incubated for 14 d under conditions described above for the pre-incubation step. The dry–rewetting disturbance was imposed at day 14 and microcosms were returned to incubation for 14 d more. Sampling of microcosms for analysis was done on day 28. The dry–rewetting disturbance consisted of air drying from the top with fan-forced air at room temperature (20–25 ∘C) during 24 h, followed by rewetting with distilled water up to 60 % WHC.
2.2 Physiological analysis
Substrate-induced respiration with p-coumaric acid and basal respiration in soil suspensions were determined with BD Oxygen Biosensor™ System microplates according to the same protocol and data processing details described in a previous study (Allegrini et al., 2017).
2.3 DNA-based analysis
2.3.1 DNA extraction and quantitation
The commercial kit PowerSoil™ DNA Isolation kit (MoBio, Inc., Carlsbad, CA) was used for DNA extraction from soil samples according to manufacturer instructions. DNA was quantified using the QuantiFluor dsDNA kit in a Quantus fluorometer (Promega Madison, WI).
2.3.2 Quantification of indicator genes
Quantification of the 16 rRNA gene, amoA gene of AOB (amoAAOB) and amoA of AOA (amoAAOA) was conducted by quantitative real-time PCR (qPCR) using the protocols described in Allegrini et al. (2015) and Zabaloy et al. (2016, 2017), respectively. For Actinobacteria the pair of primers S–P–Acti–1154–a–S–19 ∕ S–P–Acti–1339–a–A–18 was used (Pfeiffer et al., 2014). The composition of the master mix in the latter case was as follows: 7.5 µL of PCR iTaq Universal SYBR Green Supermix (2×; Bio-Rad Laboratories), 0.3 µL of each primer (stocks 10 µM, Invitrogen), and 1 µL of DNA (1–10 ng µL−1) and ultrapure water to 15 µL. The amplification programme was as follows: pre-incubation (95 ∘C, 5 min, 1 cycle), amplification (95 ∘C 15 s, 59 ∘C 30 s, 72 ∘C 45 s, 35 cycles), followed by melting curve analysis (65–95 ∘C). Decimal dilutions of a plasmid harbouring one copy of the 16S rRNA gene of Streptomyces albus DSM 40313 were used as standard (serial 10−1 dilutions to obtain between 4.97×106 and 4.97×102 copies). All amplifications were conducted in the ABI 7500 Real Time System (Applied Biosystems, Foster City, CA).
The abundance values of these genes were used as surrogates of population sizes, although no attempt was made to convert copies into cell numbers to avoid introducing errors (e.g. errors related to an unknown number of operons per cell in mixed bacterial communities) (Zabaloy et al., 2017; Ouyang et al., 2016). The efficiencies of qPCR assays were 84.1 % (amoAAOB), 78.57 % (amoAAOA), 91.07 % (total bacteria 16S rRNA) and 93.67 % (Actinobacteria 16S rRNA); and R2 values were ≥0.99 in all assays.
2.3.3 Denaturing gradient gel electrophoresis of AOB
The amplification of amoAAOB with amoA–1F ∕ amoA–2R primers (Rotthauwe et al., 1997) and the DGGE analysis of PCR products were conducted according to previously reported protocols (Allegrini et al., 2017). Digital gel images were processed with Software Gel Compare II™ v4.6 (Applied Maths). After optimization of gel properties normalization was conducted using amplicons of Nitrosomonas europaea and uncultured bacteria 5–A51 (accession number KJ643949 in GenBank) as internal reference positions (GelCompar II™ v4.6, Software Manual).
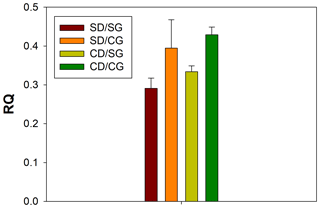
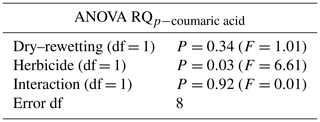
Figure 1Respiratory quotient (RQ) values. The four treatments are indicated in different colours. Error bars indicate the standard error of the mean (n=3). SD/SG: no dry–rewetting disturbance/no herbicide; SD/CG: no dry–rewetting disturbance/herbicide; CD/SG: dry–rewetting disturbance/no herbicide; CD/CG: dry–rewetting disturbance/herbicide.
2.4 Statistical analysis
RQ values were analyzed using a two-way ANOVA at a 5 % significance level using R Statistical Software v3.5.0 (R Development Core team). The copy numbers of genes (log10 copies µg−1 DNA) were analyzed in the same way. In all cases, normality and homoscedasticity were verified with Shapiro–Wilks and Levene tests, respectively (α=0.05).
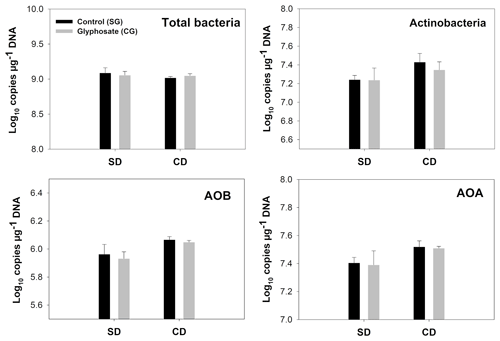
Figure 2Copy number of indicator genes for total bacteria, Actinobacteria, AOB and AOA. Error bars indicate the standard error of the mean (n=3). SD/SG: no dry–rewetting disturbance/no herbicide; SD/CG: no dry–rewetting disturbance/herbicide; CD/SG: dry–rewetting disturbance/no herbicide; CD/CG: dry–rewetting disturbance/herbicide.
Denaturing gradient gel electrophoresis fingerprints were analyzed with the software GelCompar II™ v4.6 (Applied Maths, Kortrijk, Belgium) through cluster analysis using a Pearson correlation coefficient (r) and the unweighted pair group method with arithmetic mean (UPGMA) algorithm. Cophenetic correlation coefficients were calculated in each branch and the root to determine the quality of the dendrogram. Clusters were defined at an 80 % similarity level (cut-off) and the 100 % internal stability of them (group separation assessment) was verified in GelCompar II using the statistical method of jackknife resampling with average similarities (GelCompar II™ v4.6, Software Manual).
3.1 Respiratory responses
The mean RQ values for the different treatments are indicated in Fig. 1. According to two-way ANOVA (Table 1), no interaction was observed between factors (P>0.05). Thus, the main effects were considered. No statistical significance was observed for the main effect of dry–rewetting. Conversely, herbicide showed a significant effect (P<0.05) with a higher RQ value in CG microcosms relative to the untreated microcosms (SG).
Table 2Two-way ANOVA of copy numbers for different indicator genes. The P values indicated for the main effects of herbicide and of dry–rewetting disturbances correspond to the model without interaction as no significance (P>0.05) was observed for this term. df: degrees of freedom.
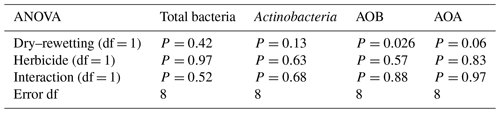
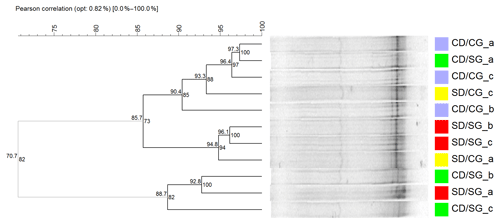
Figure 3Cluster analysis of DGGE profiles of AOB. The dendrogram was obtained using Pearson–UPGMA analysis of densitometric profiles. Treatments are indicated in different colours. Lower-case letters indicate replicates within treatments. In each node, the left number indicates the similarity value (r×100), while the right number is the cophenetic correlation coefficient. Grey branches indicate clusters with 100 % internal stability according to the jackknife method, defined at 80 % similarity value. SD/SG: no dry–rewetting disturbance/no herbicide; SD/CG: no dry–rewetting disturbance/herbicide; CD/SG: dry–rewetting disturbance/no herbicide; CD/CG: dry–rewetting disturbance/herbicide.
Table 3Copy number (copies µg−1 DNA) of the indicator genes assessed for the different microbial groups. SD/SG: no dry–rewetting/no herbicide; SD/CG: no dry–rewetting disturbance/herbicide; CD/SG: dry–rewetting disturbance/no herbicide; CD/CG: dry–rewetting disturbance/herbicide.

∗ Copy number of microcosms with (CD) or without (SD) dry–rewetting disturbance averaged through all levels of the herbicide factor.
3.2 DNA-based analysis
3.2.1 Quantification of indicator genes
For all the indicator genes, the results of two-way ANOVA (Table 2) indicated no statistical significance of the herbicide main effect as well as no interaction, while a significant dry–rewetting effect was detected only for AOB (P<0.05). Mean copy numbers for each treatment and each gene are shown in Fig. 2 and Table 3. The abundance of amoAAOB (averaged for both levels of the herbicide factor) was 1.27-fold higher in microcosms with dry–rewetting desiccation (CD) than in undisturbed (SD) microcosms (Table 3).
3.2.2 DGGE of ammonia-oxidizing bacteria
DGGE profiles showed few bands and high similarity values (Pearson coefficients) among replicates of the four treatments, with no separation in four treatment clusters. Similarly, no obvious separation was observed between microcosms with (CD) and without (SD) dry–rewetting or between glyphosate–treated (CG) and untreated microcosms (SG). At an 80 % similarity level (cut–off), a separation in two clusters was observed (Fig. 3, grey branches). In one of them, we observed two replicates of CD/SG treatment. In the second cluster the three replicates of CD/CG treatment clustered together with microcosms in which no dry–rewetting was applied (SD).
In this study we evaluated whether an acute in vitro glyphosate application on a soil with a long history of application of GBHs modulates the response of the microbial communities to the following dry–rewetting disturbance.
We hypothesized that if no PICT was observed in the studied soil after long exposure in the field (Allegrini et al., 2015), a single glyphosate application to microcosms would have no effect on the structure of the microbial community, as the probability of changing to an alternative state is more likely in response to a press disturbance (chronic exposure) than to a pulse disturbance (Shade et al., 2012). These changes in microbial communities associated with greater tolerance to a pesticide might, at the same time, conceal a higher sensitivity in the response to other perturbations (a “cost of tolerance”; Clements and Rohr, 2009). Thus, for the soil assessed in this study, we expected no conditioning effect in the sensitivity to a secondary perturbation by the presence/absence of a previous acute glyphosate exposure. This hypothesis was confirmed by our results: no interaction was observed between herbicide and dry–rewetting in an acute exposure to both perturbations with a physiological indicator (Table 1) and with DNA-based methods (Table 2). The non-significant interaction observed for Actinobacteria (Table 2) indicates that one of the main characteristics of this microbial group, the high tolerance to desiccation (Evans and Wallestein, 2011), is not conditioned by the previous exposure to a single application of a GBH, even when negative effects of GBHs on this phylum have been reported (Barriuso et al., 2010). For amoA, the absence of interaction is also a relevant observation considering that AOB are particularly sensitive to pesticides and also to water availability (Franzluebbers et al., 1994; Hastings et al., 2000; Gleeson et al., 2010). Thus, our results suggest that the sensitivity expected to each perturbation alone does not necessarily result in a synergic effect when combined.
Ammonia-oxidizing archaea were more abundant than AOB for all treatments. Also, they were clearly differentiated from AOB as no significant dry–rewetting effect was observed (Table 2). This observation is consistent with the results of Gleeson et al. (2010), who reported that AOB are more responsive to water availability than AOA. The statistical significance of the dry–rewetting main effect on the abundance of AOB indicates that the microbial community of the soil assessed in this study is particularly sensitive to the perturbation. Conversely, the abundance of AOB seems to be less sensitive to GBH exposure (no significance detected for this factor), supporting previous results with the same soil and the same herbicide formulation in which no effects of repeated applications were detected on absolute abundance (up to three applications) (Allegrini et al., 2017). As indicated in Tables 2 and 3, the dry–rewetting perturbation enhanced the abundance of amoAAOB relative to the untreated microcosms (SD). Most gram negative bacteria are affected by a rapid rewetting after desiccation events, and a recovery to the initial abundance values has been reported for AOB at 18 days after rewetting (Hastings et al., 2000). At the functional level (nitrification rate), Fierer and Schimel (2002) found a significant increase in the activity of autotrophic nitrifying communities after several dry–rewetting cycles, in agreement with the higher abundance that we observed for amoAAOB and with a correlation between amoA copy number and nitrification potential observed in different soils (Rudisill et al., 2016; Zabaloy et al., 2017).
The low number of bands observed in the DGGE profiles of amoAAOB amplicons suggests a low richness of AOB in the studied soil. This result is in agreement with a previous biogeographic study which reported a low diversity of amoA sequences in soil AOB communities, with most of them in the Nitrosospira lineages (Fierer et al., 2009). More recently, a microcosm study with a loam sandy soil from the Pampas region observed low diversity in the AOB community with DGGE (Zabaloy et al., 2017). An obvious separation among DGGE profiles of microcosms with and without dry–rewetting was not observed, indicating no effects of this perturbation on the community structure of AOB. Thus, even though qPCR indicated an increase in the abundance of amoAAOB sequences, the profiling (fingerprinting) of the community structure did not show the same sensitivity to the dry–rewetting disturbance (Fig. 3).
The separation observed at the 80 % similarity level (Fig. 3) between two replicates of CD/SG treatment and the three replicates CD/CG could be indicating an interaction as no comparable separation was detected between SD/SG and SD/CG. However, more evidence is still necessary to determine whether or not there is a significant interaction effect on the structure of AOB. Amplicon sequencing of amoAAOB and beta diversity analysis could provide substantially more information in this regard.
In conclusion, our study demonstrates that acute exposure to a GBH does not have a conditioning effect on the response of microbial communities to a secondary disturbance (dry–rewetting) in a soil with chronic exposure to GBHs. In a global context of extended use of GBHs and climate models which predict an intensification of hydrological cycles, our results are particularly relevant for the sustainability of soils in rainfed agriculture, where dry–rewetting cycles and GBH applications are expected to occur simultaneously. To obtain more evidence supporting our conclusion, future studies should assess the effects of several dry–rewetting cycles and in different soil types.
Data are available from the 4TU.ResearchData database (Allegrini, 2020; https://doi.org/10.4121/uuid:a86ce94c-1b3d-447a-8652-b2e2d0a72187).
MA, MCZ and EG designed the experiment. MA and MCZ are credited for methodology, investigation and manuscript review and editing. MA conducted formal analysis and wrote the original draft. Project administration, resources and funding acquisition were conducted by EG and MCZ.
The authors declare that they have no conflict of interest.
We thank technician Ana María Zamponi (CONICET) and trainee student Dennis Alexander Tebbe (Carl von Ossietzky Universität Oldenburg, Germany) for assistance with real-time PCR analyses.
This research has been supported by the Argentinean National Agency for Scientific and Technological Promotion (ANPCyT) (grant nos. BID-PICT 2011-0736 and BID-PICT 2012-0122).
This paper was edited by Jeanette Whitaker and reviewed by two anonymous referees.
Anderson, T. H.: Microbial eco–physiological indicators to assess soil quality, Agr. Ecosyst. Environ., 98, 285–293, https://doi.org/10.1016/S0167-8809(03)00088-4, 2003.
Allegrini, M.: Data accompanying the research on microbial response to acute dry-rewetting and glyphosate perturbations, 4TU.Centre for Research Data, Dataset, https://doi.org/10.4121/uuid:a86ce94c-1b3d-447a-8652-b2e2d0a72187, 2020.
Allegrini, M., Zabaloy, M. C., and Gómez, E.: Ecotoxicological assessment of soil microbial community tolerance to glyphosate, Sci. Total Environ., 533, 60–68, https://doi.org/10.1016/j.scitotenv.2015.06.096, 2015.
Allegrini, M., Gómez, E., and Zabaloy, M. C.: Repeated glyphosate exposure induces shifts in nitrifying communities and metabolism of phenylpropanoids, Soil Biol. Biochem., 105, 206–215, https://doi.org/10.1016/j.soilbio.2016.11.024, 2017.
Barriuso, J., Marín, S., and Mellado, R. P.: Effect of the herbicide glyphosate on glyphosate-tolerant maize rhizobacterial communities: a comparison with pre-emergency applied herbicide consisting of a combination of acetochlor and terbuthylazine, Environ. Microb., 12, 1021–1030, https://doi.org/10.1111/j.1462-2920.2009.02146.x, 2010.
Bustamante, M., Verdejo, V., Zúñiga, C., Espinosa, F., Orlando, J., and Carú, M.: Comparison of water availability effect on ammonia–oxidizing bacteria and archaea in microcosms of a Chilean semiarid soil, Front. Microbiol. 3, 282, https://doi.org/10.3389/fmicb.2012.00282, 2012.
Cerdeira, A. L. and Duke, S. O.: The current status and environmental impacts of glyphosate–resistant crops. A review, J. Environ. Qual., 35, 1633–1658, https://doi.org/10.2134/jeq2005.0378, 2006.
Clements, W. H. and Rohr, J. R.: Community responses to contaminants: using basic ecological principles to predict ecotoxicological effects, Environ. Toxicol. Chem., 28, 1789–1800, https://doi.org/10.1897/09-140.1., 2009.
Evans, S. E. and Wallenstein, M. D.: Soil microbial community response to drying and rewetting stress: does historical precipitation regime matter?, Biogeochemistry, 109, 101–116, https://doi.org/10.1007/s10533-011-9638-3, 2011.
EFSA Panel on Plant Protection Products and their Residues: Scientific Opinion Addressing the State of the Science on Risk Assessment of Plant Protection Products for In–soil Organisms, EFSA Journal, European Food Safety Authority, Parma, available at: https://efsa.onlinelibrary.wiley.com (last access: January 2020), 2016.
Fierer, N. and Schimel, J.: Effects of drying–rewetting frequency on soil carbon and nitrogen transformations, Soil Biol. Biochem., 34, 777–787, https://doi.org/10.1016/S0038-0717(02)00007-X, 2002.
Fierer, N., Schimel, J. P., and Holden, P. A.: Influence of Drying–Rewetting Frequency on Soil Bacterial Community Structure, Microb. Ecol., 45, 63–71, https://doi.org/10.1007/s00248-002-1007-2, 2003.
Fierer, N., Carney, K. M., Horner-Devine, M. C., and Megonigal, J. P.: The biogeography of ammonia–oxidizing bacterial communities in soil, Microb. Ecol., 58, 435–445, https://doi.org/10.1007/s00248-009-9517-9, 2009.
Franzluebbers, A. J., Hons, F. M., and Zuberer, D. A.: Long-term changes in soil carbon and nitrogen pools in wheat management systems, Soil Sci. Soc. Am. J., 58, 1639–1645, https://doi.org/10.2136/sssaj1994.03615995005800060009x, 1994.
GelCompar II™ v4.6 Software Manual: Applied Maths, Kortrijk Belgium, available at: http://www.applied-maths.com/, last access: May 2016.
Gleeson, D. B., Herrmann, A. M., Livesley, S. J., and Murphy D. V.: Influence of water potential on nitrification and structure of nitrifying bacterial communities in semiarid soils, App. Soil Ecol., 40, 189–194, https://doi.org/10.1016/j.apsoil.2008.02.005, 2008.
Gleeson, D. B., Müller, C., Banerjee, S., Ma, W., Siciliano, S. D., and Murphy, D. V.: Response of ammonia oxidizing archaea and bacteria to changing water filled pore space, Soil Biol. Biochem., 42, 1888–1891, https://doi.org/10.1016/j.soilbio.2010.06.020, 2010.
Griffiths, R. I., Whiteley, A. S., O'Donnell, A. G., and Bailey, M. J.: Physiological and community responses of established grassland bacterial populations to water stress, App. Environ. Microbiol., 69, 6961–6968, https://doi.org/10.1128/AEM.69.12.6961-6968.2003, 2003.
Haney, R. L., Senseman, S. A., Hons, F. M., and Zuberer, D. A.: Effect of glyphosate on soil microbial activity and biomass, Weed Sci. 48, 89–93, https://doi.org/10.1614/0043-1745(2000)048[0089:EOGOSM]2.0.CO;2, 2000.
Hastings, R. C., Butler, C., Singleton, I., Saunders, J. R., and McCarthy, A. J.: Analysis of ammonia–oxidizing bacteria populations in acid forest soil during conditions of moisture limitation, Lett. Appl. Microbiol., 30, 14–18, https://doi.org/10.1046/j.1472--765x.2000.00630.x., 2000.
Huntington, T. G.: Evidence for intensification of the global water cycle: review and synthesis, J. Hydrol., 319, 83–95, https://doi.org/10.1016/j.jhydrol.2005.07.003, 2006.
Ouyang, Y., Norton, J. M., Stark, J. M., Reeve, J. R., and Habteselassie, M. Y.: Ammonia–oxidizing bacteria are more responsive than archaea to nitrogen source in an agricultural soil, Soil Biol. Biochem., 96, 4–15, https://doi.org/10.1016/j.soilbio.2016.01.012, 2016.
Pfeiffer, S., Pastar, M., Mitter, B., Lippert, K., Hackl, E., Lojan, P., Oswald, A., and Sessitsch, A.: Improved group–specific primers based on the full SILVA 16S rRNA gene reference database, Environ. Microbiol., 16, 2389–2407, https://doi.org/10.1111/1462-2920.12350, 2014.
Ratcliff, A. W., Busse, M. D., and Shestak, C. J.: Changes in microbial community structure following herbicide (glyphosate) additions to forest soils, Appl. Soil Ecol., 34, 114–124, https://doi.org/10.1016/j.apsoil.2006.03.002, 2006.
Rotthauwe, J. H., Witzel, K. P., and Liesack, W.: The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations, Appl. Environ. Microbiol., 63, 4704–4712, 1997.
Rudisill, M. A., Turco, R. F., and Hoagland, L. A.: Fertility practices and rhizosphere effects alter ammonia oxidizer community structure and potential nitrification activity in pepper production soils, Appl. Soil Ecol., 99, 70–77, https://doi.org/10.1016/j.apsoil.2015.10.011, 2016.
Shade, A., Peter, H., Allison, S. D., Baho, D. L., Berga, M., Bürgmann, H., Huber, D. H., Langenheder, S., Lennon, J. T., Martiny, J. B. H., Matulich, K. L., Schmidt, T. M., and Handelsman, J.: Fundamentals of microbial community resistance and resilience, Front. Microbiol. 3, 417, https://doi.org/10.3389/fmicb.2012.00417, 2012.
Tilman, D., Cassman, K. G., Matson, P. A., Naylor, R., and Polasky, S.: Agricultural sustainability and intensive production practices, Nature, 418, 671–677, https://doi.org/10.1038/nature01014, 2002.
Zabaloy, M. C., Carné, I., Viassolo, R., Gómez, M. A., and Gomez, E.: Soil ecotoxicity assessment of glyphosate use under field conditions: microbial activity and community structure of Eubacteria and ammonia-oxidising bacteria, Pest Manage. Sci., 72, 684–691, https://doi.org/10.1002/ps.4037, 2016.
Zabaloy, M. C., Allegrini, M., Tebbe, D. A, Schuster, K., and Gómez, E.: Nitrifying bacteria and archaea withstanding glyphosate in fertilized soil microcosms, Appl. Soil Ecol., 117, 88–95, https://doi.org/10.1016/j.apsoil.2017.04.012, 2017.
Zhang, M., Wang, W., Tang, L., Heenan, M., and Xu, Z.: Effects of nitrification inhibitor and herbicides on nitrification, nitrite and nitrate consumptions and nitrous oxide emission in an Australian sugarcane soil, Biol. Fert. Soils, 54, 697–706, https://doi.org/10.1007/s00374-018-1293-6, 2018.