the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Potential of natural language processing for metadata extraction from environmental scientific publications
Guillaume Blanchy
Lukas Albrecht
John Koestel
Sarah Garré
Summarizing information from large bodies of scientific literature is an essential but work-intensive task. This is especially true in environmental studies where multiple factors (e.g., soil, climate, vegetation) can contribute to the effects observed. Meta-analyses, studies that quantitatively summarize findings of a large body of literature, rely on manually curated databases built upon primary publications. However, given the increasing amount of literature, this manual work is likely to require more and more effort in the future. Natural language processing (NLP) facilitates this task, but it is not clear yet to which extent the extraction process is reliable or complete. In this work, we explore three NLP techniques that can help support this task: topic modeling, tailored regular expressions and the shortest dependency path method. We apply these techniques in a practical and reproducible workflow on two corpora of documents: the Open Tension-disk Infiltrometer Meta-database (OTIM) and the Meta corpus. The OTIM corpus contains the source publications of the entries of the OTIM database of near-saturated hydraulic conductivity from tension-disk infiltrometer measurements (https://github.com/climasoma/otim-db, last access: 1 March 2023). The Meta corpus is constituted of all primary studies from 36 selected meta-analyses on the impact of agricultural practices on sustainable water management in Europe. As a first step of our practical workflow, we identified different topics from the individual source publications of the Meta corpus using topic modeling. This enabled us to distinguish well-researched topics (e.g., conventional tillage, cover crops), where meta-analysis would be useful, from neglected topics (e.g., effect of irrigation on soil properties), showing potential knowledge gaps. Then, we used tailored regular expressions to extract coordinates, soil texture, soil type, rainfall, disk diameter and tensions from the OTIM corpus to build a quantitative database. We were able to retrieve the respective information with 56 % up to 100 % of all relevant information (recall) and with a precision between 83 % and 100 %. Finally, we extracted relationships between a set of drivers corresponding to different soil management practices or amendments (e.g., “biochar”, “zero tillage”) and target variables (e.g., “soil aggregate”, “hydraulic conductivity”, “crop yield”) from the source publications' abstracts of the Meta corpus using the shortest dependency path between them. These relationships were further classified according to positive, negative or absent correlations between the driver and the target variable. This quickly provided an overview of the different driver–variable relationships and their abundance for an entire body of literature. Overall, we found that all three tested NLP techniques were able to support evidence synthesis tasks. While human supervision remains essential, NLP methods have the potential to support automated evidence synthesis which can be continuously updated as new publications become available.
- Article
(3242 KB) - Full-text XML
- BibTeX
- EndNote
The effect of agricultural practices on agroecosystems is highly dependent upon other environmental factors such as climate and soil. In this context, summarizing information from scientific literature while extracting relevant environmental variables is important to establish conclusions that are specific to pedo-climatic information. This synthesis is essential to provide recommendations for soil management adaptations that are adequate for local conditions, both today and in the future. Efforts to synthesize context-specific evidence through meta-analysis or reviews currently require a lot of manual work to extract specific information from papers. This effort scales with the number of publications, which makes it more difficult to collate exhaustive meta-databases from the literature. In the meantime, the use of automated methods to analyze unstructured information (like text in a scientific publication) has been developing during recent years and has demonstrated potential to support evidence synthesis (Haddaway et al., 2020). Natural language processing (NLP) is one of them.
In their 2019 review on the advances of the technique, Hirschberg and Manning explained that “natural language processing employs computational techniques for the purpose of learning, understanding and producing human language content”. This definition is quite broad as NLP encompasses several considerably different techniques, like machine translation, information extraction or natural language understanding. Nadkarni et al. (2011) and Hirschberg and Manning (2015) provide a good overview on this field of research and how it originally developed. For applications to scientific publications, Nasar et al. (2018) reviewed different NLP techniques (information extraction, recommender systems, classification and clustering, and summarizations). However, an important limitation of supervised NLP techniques is that they require labels that need to be manually produced to train the model. Hence, humans are still needed for evidence synthesis but can certainly receive great support from existing NLP techniques.
NLP methods are most widely used in medical research. The development of electronic health records significantly facilitated the application of automatic methods to extract information. For instance, information extraction techniques were used to identify adverse reactions to drugs, identify patients with certain illnesses which were not discovered yet at the time or link genes with their respective expression (Wang et al., 2018). A specific example is given by Tao et al. (2017), who used word embedding and controlled random fields to extract prescriptions from discharge summaries. Wang et al. (2018) provide an extensive review of the use of NLP for the medical context.
The rise of open-source software tools such as NLTK (Loper and Bird, 2002) and SpaCy (Honnibal and Montani, 2017) together with the increase in digitally available information has fostered the way for NLP applications towards other scientific communities. For example, SpaCy is able to recognize the nature of words in a sentence and their dependence on other words using a combination of rule-based and statistical methods. Building on that, there are open-source software tools that aim at automatically extracting information. A very popular tool is the OpenIE framework of the Stanford group (Angeli et al., 2015), included in the Stanford coreNLP package (Manning et al., 2014). Niklaus et al. (2018) present a review of open-source information extraction codes. All these tools greatly support novel implementations of NLP applications as they reduce the knowledge required for new users to start using NLP techniques.
In the context of evidence synthesis, several NLP methods can be useful. Topic modeling can help to identify common themes in a corpus of publications or classify publications by subject. In addition to selecting publications to be reviewed in the evidence synthesis, topic modeling also gives an overview of the number of publications per topic and helps to identify knowledge gaps. Regular expressions search the text for a pattern of predefined numbers and words. They have a high precision but only find what they are designed to find. This means the user already needs a lot of knowledge on the exact words/terms that should be found. They can be augmented by including syntaxic information such as the nature (noun, adjective, adverb, etc.) and function (verb, subject, etc.) of a word. Complemented with dictionaries that contain lists of specific words (e.g., World Reference Base soil groups), it can be a powerful method. More advanced NLP techniques aim at transforming words into numerical representations that can be further processed by numerical machine learning algorithms. For instance, word embeddings are vectors which encode information about a word and its linguistic relationships in the context it is found. They are derived from the corpus of available documents. One group of advanced machine learning techniques that convert text to a numerical representation are transformer networks such as BERT (Koroteev 2021). BERT transformers are trained on specific corpora. For instance bioBERT is tailored to the medical context (Lee et al., 2020).
In contrast to the medical context, fewer studies applied NLP methods to soil sciences. Padarian et al. (2020) used topic modeling in their review of the use of machine learning in soil sciences. Furey et al. (2019) presented NLP methods to extract pedological information from soil survey descriptions. Padarian and Fuentes (2019) used word embedding. They were able to establish relationships between soil types and soil or site properties through principal component analysis. For instance, “Vertisols” were associated with “cracks” or “Andosols” with “volcanoes” as their embeddings were similar.
The aim of this study is not to demonstrate the latest and most advanced NLP techniques. Rather, it presents practical workflows to apply NLP techniques to scientific publication in soil science to support different evidence synthesis steps: topic classification, knowledge gap identification and database building. Our study aims to demonstrate the potential and practical limitations of several NLP techniques through examples of evidence synthesis for soil science. In this regard, we put special emphasis on the methodology used and its ability to recover information, rather than analyzing and interpreting the extracted data itself. We redirect the reader to chapters 1–3 in Garré et al. (2022) for detailed interpretation of the evidence synthesis.
The objectives of this paper are (1) to demonstrate the potential of natural language processing as for the collection of structured information from scientific publications, (2) to illustrate the ability of topic classification to identify “popular” and less investigated topics, and (3) to assess the ability of natural language processing to extract relationships between a given driver (tillage, cover crops, amendment, etc.) and soil variables (hydraulic conductivity, aggregate stability, etc.) based on publication abstracts.
2.1 Text corpora
This work used two corpora (sets of texts) which are referred to in the following as the OTIM and the Meta corpus. The OTIM corpus was related to OTIM-DB (https://doi.org/10.20387/bonares-q9b3-z989, EJP SOIL – CLIMASOMA, 2022, chapter 4) which is a meta-database extending the one analyzed in Jarvis et al. (2013) and Jorda et al. (2015). OTIM-DB contains information about the near-saturated hydraulic conductivity obtained from the tension-disk infiltrometer between 0 and −10 cm tension. OTIM-DB also includes metadata on the soil (texture, bulk density, organic carbon content, World Reference Base (WRB) classification); 23 climatic variables that were assigned based on the coordinates of the measurement locations, among them annual mean temperature and precipitation; methodological setup (disk diameter, method with which infiltration data are converted to hydraulic conductivity, month of measurement); and land management practices (land use, tillage, cover crops, crop rotation, irrigation, compaction). All data in OTIM-DB were manually extracted by researchers from 172 source publications. The collected data were then cross-checked by another researcher to catch typos and misinterpretations of the published information. The OTIM corpus consisted of the entire texts of the 172 source publications used in OTIM-DB.
In contrast, the Meta corpus contained only abstracts, namely, the ones of the primary studies included in the meta-analyses by EJP SOIL – CLIMASOMA (2022, chapter 1), which investigated how soil hydraulic properties are influenced by soil management practices. This Meta corpus contained 1469 publications. By number of publications, it was substantially larger than the OTIM corpus. The information given in the Meta corpus was not available in a meta-database. Therefore, the validation step had to be carried out by manually extracting information from a subset of the abstracts in this corpus. The references for both the OTIM and Meta corpora are available on the GitHub repository of this project (https://doi.org/10.5281/zenodo.7687794).
2.2 Extracting plain text from the PDF
For both corpora, all publications were retrieved as PDF files. The software “pdftotext” (https://www.xpdfreader.com/pdftotext-man.html, last access: 1 March 2023) was used to extract the text from these PDFs. The text extraction worked well apart from one exception where the extracted text contained alternating sentences from two different text columns, making it unsuited for NLP. Other two-column publications were correctly extracted. Other methods were tested, such as the use of the Python package PyPDF2 or the use of the framework pdf.js, but did not provide better results than pdftotext. The difficulty of this conversion lies in the format of the PDF itself that locates words in reference to the page and does not preserve information on which words belong to individual sentences or paragraphs. Recovery methods (such as in pdf.js or pdftotext) use the distance between words to infer if they belong to the same sentence and detect paragraphs. This makes extracting text from PDF harder for algorithms and is clearly a major drawback of this format. In addition, text boxes and figures can span multiple text columns and make the conversion difficult (e.g., the figure caption intercalated in the middle of the text). This led Ramakrishnan et al. (2012) to develop LA-PDFText, a layout-aware PDF-to-text translator designed for scientific publications (which was not used in this study due to time restriction). The addition of a hidden machine-friendly text layer in the PDF itself or the use of the full-text HTML version can possibly alleviate this issue. Another limitation of PDF is that tables are encoded as a series of vertical/horizontal lines placed at a given position. When converting the PDF to text, only streams of numbers can be retrieved. Rebuilding the tables based on the regularity of the spacing between these numbers is possible in some cases (e.g., Rastan et al., 2019), but nevertheless understanding what these numbers represent based solely on the headers is for now out of reach of the NLP algorithm. For this issue too, HTML format has an advantage as tables are encoded in the HTML or provided as separate .xlsx or .csv files, hence enabling easier information extraction.
However, because online full-text HTML pages were not available for all documents (mostly older publications) and PDF remains the most widespread format for exchanging scientific publication, we decided to pursue the analysis with PDF. From the extracted full-text pages, abstract and reference sections were removed, and only the body of the text was used to form the documents for each corpus.
Several NLP techniques were applied. Table 1 summarizes which techniques were applied to which corpus.
2.3 Topic modeling
Topic modeling creates topics from a corpus by comparing the similarity of the words between documents. We extracted all bigrams (two consecutive words) that occurred more than 20 times for each document in the Meta corpus. Each document was then represented by its bigrams. We found that using bigrams instead of single words helps to obtain more coherent topics. This is intuitively explained by considering that the bigrams “cover crops” and “conventional tillage” are more informative than “cover”, “crops”, “conventional” and “tillage” alone. Next, we removed all bigrams that appeared in less than 20 documents and more than 50 % of all documents to avoid too specific or too common bigrams. Note that bigrams are usually added to monograms. However, in our case, we found that bigrams alone (e.g., “conventional tillage”, “cover crops”) led to more coherent topics than when combined with monograms (e.g., topics around “tillage”, “crops”, “water”). Topics were then created to be as coherent as possible using the latent Dirichlet allocation (LDA) algorithm. A number of different coherence metrics exist (Röder et al., 2015). In this work, we used the LDA implementation of the gensim library (v4.1.2) with the CV coherence metric. This CV coherence metric is a combination of a normalized pointwise mutual information coherence measure, cosine vector similarity and a Boolean sliding window of size 110 words as defined in Röder et al. (2015). The metric ranges from 0 (not coherent at all) to 1 (fully coherent). To define the optimal number of topics to be modeled, we iteratively increased the number of topics from 2 to 20 and looked at the coherence. Based on this optimal number of topics, the composition of each topic was further analyzed using the figures generated by the pyLDAvis package (v3.3.1), which is based upon the work of Sievert and Shirley (2014).
2.4 Rule-based extraction
Regular expressions are predefined patterns that can include text, numbers
and symbols. For instance, disk diameters of tension-disk infiltrometers are
extracted from a text using the regular expression search term “(\d+\.\d)cm\s diameter”, which will
retrieve text passages like “5.4 cm diameter”. In this regular expression,
\d denotes a digit, \s a space, and \d+ one or more digits, and parentheses are used to enclose the group we want to
extract. Regular expressions are a widely used rule-based extraction tool in
computer science. They have a high precision, but their complexity can
quickly increase for more complex patterns. Figure 1 provides examples of
regular expressions used in our study. It can be observed that regular
expressions for geographic coordinates are quite complex as they need to
account for different notations such as decimal format (24.534∘ N) or
degree–minute–second format (24∘4′23.03′′ N) in the case of
latitudes. In contrast, specific well-defined terms such as World Reference
Base (WRB) soil types are more easily retrieved as their wording is always
unique in the text. Soil textures are likewise easy to extract but less
well-defined as, for example, WRB soil types. Often, terms used to describe
the soil texture of an investigated field site are also used to refer to
general cases or unrelated field sites in the same text. This makes it more
challenging to automatically extract information on the investigated site
using regular expressions. To complicate matters, soil textures are not
always given in the USDA (United States Department of Agriculture)
classification system, which can be regarded as a standard. For the sake of
simplicity, we did not attempt to identify the texture classification system
but treated all textural information as if it was using the USDA system.
When gathering information on tension-disk diameters, we must pay attention to the
length units as well as whether the radius or the diameter
was reported. In these more complex cases, we constructed the regular
expression search terms iteratively to extract the greatest amount of
information from the available papers. Regular expressions were used to
extract latitude, longitude, elevation, soil type, soil texture, annual
rainfall, disk size and tensions applied.
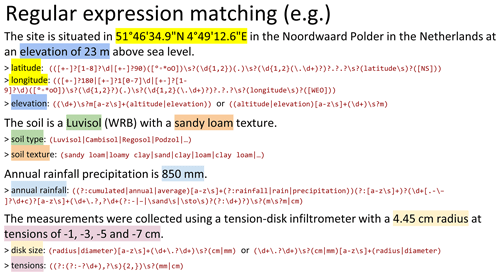
Figure 1Examples of different regular expressions used for information extraction. \d represents a digit, \s a space, . (a dot) an unspecified character, and [a–z] all lower case letters, and more generally squared brackets are used to denote the list of characters (e.g., [∘∗o0] is used to catch the symbol for degrees in latitude and longitude). A character can be “present once or absent” (\d\.\d? will match both integers and decimal numbers), “present at least once” (\d+ will match 7, 73 and 735) or “present at given number of times” (\d{1,2} will match 7 and 73 but not 735). Parentheses are used to segment capturing groups and can also contain Boolean operator such as OR denoted by | which is used to catch exact WRB soil names (Luvisol | Cambisol | etc.). Non-capturing parentheses are denoted with (?:) like for the regular expression of tensions. The content inside non-capturing parentheses will not be outputted as results of the regular expression in contrast to other parenthesis groups.
To assess the quality of the extraction, different metrics were used. They are illustrated in Fig. 2. For rule-based extraction, two tasks are required by the algorithm: selection and matching. The selection task aims to assess the ability of the algorithm to extract relevant information from the text. The matching task assesses the ability of the algorithm to extract not only the relevant information but also the correct specific information as recorded in the database used for validation. For instance, if the NLP algorithm identified “Cambisol” as the soil group for a study conducted on a Cambisol, the selection was true positive (TP) and the matching was true. If the text did not contain any WRB soil type and the NLP did not return any, both selection and matching performed well with the selection being a true negative (TN) case. Eventually, when the NLP algorithm did not find a WRB soil type but the database listed one, the selection was referred to as false negative (FN) and the matching as false. The opposite case, with a soil type found in the text but no entry in the database, was called false positive (FP) and the matching was equally false. Eventually, there were cases where the NLP algorithm retrieved incorrect information but still provided a meaningful value, e.g., if the algorithm extracted “Luvisol” as the soil type while the correct value was “Cambisol”. Then the selection task was still successful since the found term represents a WRB soil type. However, the matching task failed. Such cases were still marked as true positives but with a false matching.
Four different metrics were used to evaluate the results: the recall, the precision, the F1 score and the matching score. The recall assesses the ability of the algorithm to find all relevant words in the corpus (recall = 1). The precision assesses the ability of the algorithm to only select relevant words (precision = 1). If there were 100 soil types to be found in the corpus and the algorithm retrieved 80 words of which 40 were actually soil types, the recall was 40/100 = 0.4, and the precision was 40/80 = 0.5. The F1 score combines the recall and precision in one metric which is equal to 1 if both recall and precision were equal to 1. The recall, precision and F1 scores were used to assess the ability of the algorithm to extract relevant information from the text. Figure 2 also includes the equations for recall, precision, F1 score and matching score. Note the difference between precision and matching score: the precision expresses how many relevant words were extracted, while the matching score quantifies the fraction of words corresponding to the correct information. Considering the example above, if out of the 40 correctly selected soil types only 20 actually matched what was labeled in the document, then the matching score would be 20/100 = 0.2. Figure A1 in the Appendix gives a graphical overview of the recall and precision metrics. In addition to these metrics, a matching score was used to illustrate how many NLP-extracted values actually matched the one manually entered in the database. All rule-based extraction expressions were applied on the OTIM corpus, and the information stored in OTIM-DB was used for validation.
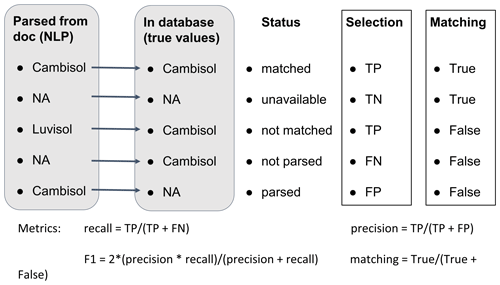
Figure 2Cases of NLP extraction results in regards to the value entered in the database (considered the correct values) for the selection task and the matching task. TP, TN, FN and FP stand for true positive, true negative, false negative and false positive, respectively. The recall, precision, F1 score and matching values served as metrics for each task.
In addition to the above extraction rules, we also identified agricultural practices mentioned in the publications and the co-occurrence of pairs of practices within the same publications. This enabled us to highlight which practices are often associated. To identify management practices in the OTIM corpus, we used the list of keywords from the Bonares Knowledge Library (https://klibrary.bonares.de/soildoc/soil-doc-search, last access: 1 March 2023). Given that several keywords can relate to the same practice, the list was further expanded by using the synonyms available from the FAO thesaurus AGROVOC (Caracciolo et al., 2013).
2.5 Relationship extractions
Relationship extraction relates drivers (agricultural practices) defined by specific key terms to specific variables (soil and site properties). In this study, examples for drivers were “tillage”, “cover crop” or “irrigation”. Among the investigated variables were “hydraulic conductivity”, “water retention” or “yield”. A list of all considered drivers and variables is given in Table A1 in the Appendix. These keywords corresponded to the keywords used in early stages of assembling the Meta corpus (EJP SOIL – CLIMASOMA, 2022, chapter 1). To allow for catching both plural and singular forms, all drivers and keywords were converted to their meaningful root: their lemma (e.g., the lemma of “residues” is “residue”).
The relationship extraction algorithm searched in the Meta corpus for sentences which contained lemmas of both drivers and variables. Each sentence was then split into words (tokenized), and each word (token) was assigned a part-of-speech (POS) tag (e.g., noun, verb, adjective). Dependencies between the tokens were also computed. Using these dependencies as links, a graph with one node per token was built. The nodes corresponding to the driver and variables were identified, and the shortest dependency path between them was computed (Fig. 3).
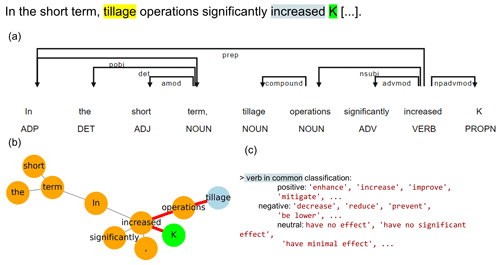
Figure 3Example of NLP extraction on a sentence. Panel (a) shows the part-of-speech (POS) tag below each token and the dependencies (arrows) to other tokens. (b) Based on these dependencies, a network graph was created, and the shortest dependency path between the driver (blue circle) and the variable (green circle) is shown in red. (c) The verb contained in the shortest dependency path was classified into positive, negative or neutral according to pre-established lists.
Table 2Examples of relationships identified and their corresponding classified labels. Note that the modifiers present in the noun chunk (e.g., “conservation tillage” or “zero tillage”) and the negation in the sentence were taken into account in the status of the relationship. Some sentences contain multiple driver–variable pairs and, hence, multiple relationships. In such cases, only one of the two was indicated in the table below (but all were considered in the code).
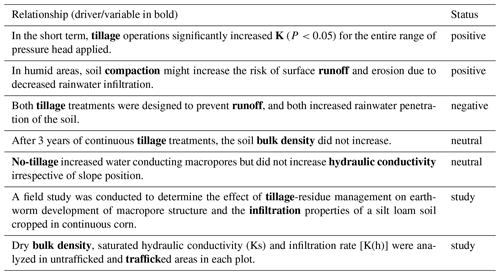
All tokens that were part of this shortest dependency path between the driver token and the variable token were kept in a list. From this list, the tokens containing the driver/variables were replaced by the noun chunk (i.e., groups of nouns and adjectives around the token) as important information can be contained in this chunk. For instance, the driver token “tillage” was replaced by its noun chunk “conventional tillage”. The list of tokens that constituted the shortest dependency path always included the main verb linking the driver and the variable token. This verb depicted a positive, negative or neutral correlation between the driver and the variable. Other modifiers such as negation marks or other modifiers that can be part of the noun chunk (e.g., “conservation” or “conventional” with the noun “tillage”) were also searched for in each sentence. In cases where a positive correlation was negated (e.g., “did not increase”, “did not have significant effect on”), the relationship was classified as neutral. Sometimes, the relationship did not relate directly to the correlation between the driver and the variable but rather mentions that this relationship was studied in the paper. Then, the status of the relationships was set to “study”. To assess the recall and precision of the technique, a subset of 129 extracted relationships was manually labeled. Table 2 offers examples of relationships classified by the algorithm.
When identifying driver and variable pairs among abstracts, the case can be encountered where one of the drivers/variables is expressed using a pronoun. This prevents keyword-based detection. The neuralcoref Python package was used to replace the pronouns by their initial form using co-references. This package uses neural networks to establish a link between the pronoun and the entity it refers to. The pronoun is then replaced by the full text corresponding to the entity. For the Meta corpus, the co-reference substitution did not enable us to increase the number of relevant sentences extracted. It turned out that the use of pronouns in the investigated abstracts was very limited. In addition, the accuracy of the co-reference substitution was not always relevant, and substitution errors were more frequent than desired. For these reasons, we left this step out of the final processing pipeline. Nevertheless, we want to stress that replacing pronouns may be very useful for other corpora. Automatic relationship extraction using OpenIE was also tried, but given the specificity of the vocabulary in the corpus of abstracts, it yielded relatively poor results.
To ensure reproducibility, all codes used in this project were written down in Jupyter Notebooks. This enabled the results to be replicated and the code to be reused for other applications. Jupyter Notebooks also enable figures and comments to be placed directly inside the document, hence helping the reader to better understand the code snippets. All notebooks used in this work are freely available on GitHub https://github.com/climasoma/nlp/ (last access: 1 March 2023).
3.1 Topic modeling
Figure 4 shows the evolution of the coherence metric with respect to the number of topics. The overall model coherence increases up to six topics then slowly starts to decrease. Note that the coherence scores can slightly change between runs as the algorithm starts from a different random seed to build the topics. Nevertheless, we observed a stagnation of the overall coherence after six topics, meaning that increasing the number of topics above six did not increase the overall coherence of the model.
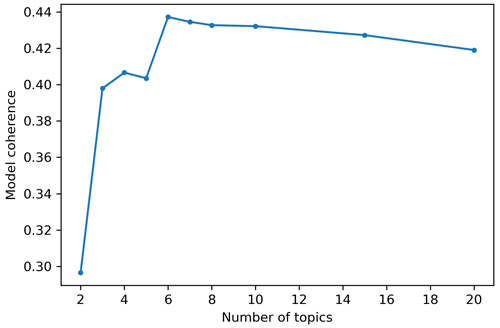
Figure 4Evolution of overall model coherence according to the number of topics chosen to train the LDA model. The coherence metric is the CV described in Röder et al. (2015), which is a combination of a normalized pointwise mutual information coherence measure, cosine vector similarity and a Boolean sliding window of size 110.
Figure 5 (left) shows the frequency of the topic in the corpus (as percentage of documents in the corpus that belong to this topic). The circles are placed according to the first two principal components based on their inter-topic distance computed using the Jensen–Shannon divergence (Lin, 1991). Topics closer to each other are more similar than topics further apart. Figure 5 (right) shows the frequency of each bigram in the topic and in the corpus. Different themes are visible from the topics: microbial biomass and aggregate (topic 1), conventional/conservation tillage (topic 2), crop residue and crop rotation (topic 3), water retention and porosity (topic 4), infiltration rate and grazing (topic 5), and cover crops (topic 6). The left part of Fig. 5 shows how topics 1 to 4 are close in contrast to topic 6 that mainly focuses on cover crops. These subtopics nicely correspond to some of the main drivers initially set in the search query string used to build the Meta corpus (EJP SOIL – CLIMASOMA, 2022, chapter 1). The topic modeling shows that bigrams such as “cover crops” have a large term frequency (blue bars), which means they are relatively frequent inside the set of documents. Bigrams such as “conservation tillage”, “aggregate stability” or “microbial biomass” are less frequent (smaller blue bars). The topic modeling also shows that terms such as “grain yield” appear in several topics (topics 2 and 3). But it is more frequent in topic 2 than topic 3 (size of the orange bars). On the contrary, bigrams such as “hairy vetch” or “winter cover” are entirely specific to topic 6 on cover crops. It should be also noted that bigrams such as “deficit irrigation”, while present in the corpus, do not appear in the top-six more relevant terms. This shows that this theme is less represented in the corpus and possibly indicates a knowledge gap around it. Another possible explanation is that, while few papers mentioned “deficit irrigation”, the inclusion of monograms such as “irrigation” might have led to the construction of a topic around irrigation techniques (where papers around “deficit irrigation” might have been found). While different runs of the LDA led to slightly different topic distributions due to the randomness internally used by the algorithm, we observed the appearance of the same coherent topics around tillage, cover crops or biochar. This highlights the fact that the LDA is not a deterministic method but a probabilistic one as the probability of a topic per document and the probability of a term per topic are iteratively optimized to maximize the coherence by the LDA. Overall, we consider that topic modeling can serve as a first tool for an exploratory analysis of the corpus content.
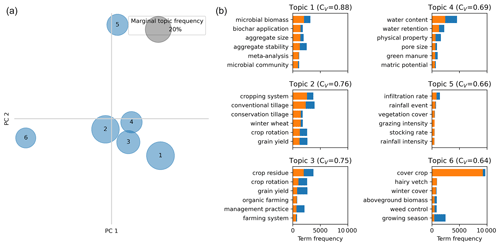
Figure 5(a) Map of topics according to the first two principal components after dimension reduction. (b) For each topic, the six more relevant bigrams inside the topic are shown. The orange bars represents the term frequency inside the topic, while the length of the full bars (orange + blue) represent the term frequency in the entire corpus. The gray circle represents the size of a topic that contains 20 % of the documents of the corpus.
3.2 Rule-based extraction
Table 3 shows the metrics relative to the different rule-based extraction techniques. Note that “n” does not always represent the number of documents in the corpus as a document can contain multiple locations for instance. Regular expressions associated with a dictionary for soil texture and soil type provide the best precision overall due to their high specificity. This clearly highlights the usefulness of the international scientific community agreeing on a common vocabulary or classification system. Soil type had the highest recall, which means that all instances of soil types mentioned in the document had been successfully extracted. Regular expression matching quantities such as “rainfall”, “disk diameter”, “tensions” or “coordinates” had lower recall than rules making use of a dictionary. Coordinates had a high precision but a lower recall as some coordinate formats could not be extracted from the text. This could be partly explained by the conversion of the symbols for degree–minute–seconds from PDF to text. As the encoding of these characters varies a lot between journals, the conversion sometimes led to “∘” converted to “O”, “∗” or “0”. Identifying all these different cases while retaining a high accuracy on more frequent cases was challenging with regular expressions.
Table 3Scores of the rule-based extraction methods. n is the number of items to be extracted. It varies as several coordinates can be provided in the same paper. The method can use only a regular expression (regex) or a combination of regular expression and dictionary (regex + dict.).
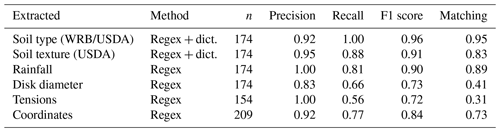
Regular expressions have to be flexible enough to accommodate the various formats found in the publications (e.g., for coordinates) but also discriminant enough to not match irrelevant items. For instance, the regular expression about soil texture catches a lot of terms related to soil texture, but not all were related to the soil texture of the actual field site. Applying regular expression on specific parts of the paper (for instance, just on the materials and methods section) could help improve the precision of the technique. Note that the regular expression algorithm itself is infallible by nature (it will always return exactly what is matched). Rather, here, we assess our ability to generate regular expression patterns that are general enough to extract information for most cases. Adjusting the regular expression to fit all edge cases encountered is, in theory, possible but will be work intensive and will not scale well with an increasing number of papers.
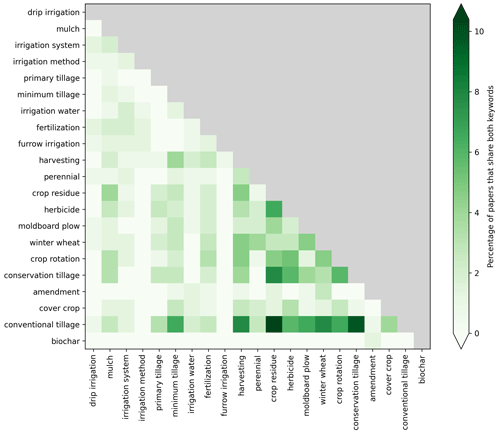
In addition to extracting specific data, general information about which management practices are investigated in the studies is also important. Figure 6 shows the co-occurrence of the detected practices inside the same document as the percentage of documents in the OTIM corpus that contains both practices. For instance, the practice of “crop residue” and “conversion tillage” is often found with documents that contain “conventional tillage”. This can be put in parallel with the topic modeling where these two bigrams were often associated. The term “herbicide” is also often mentioned with documents containing “crop residue”. Given the small size of the chosen corpus, the co-occurrences need to be interpreted in connection to experimental sites chosen for tension-infiltrometer measurements and hence provide an overview of which practices have been most studied with tension-disk infiltrometers.
3.3 Relationship extraction
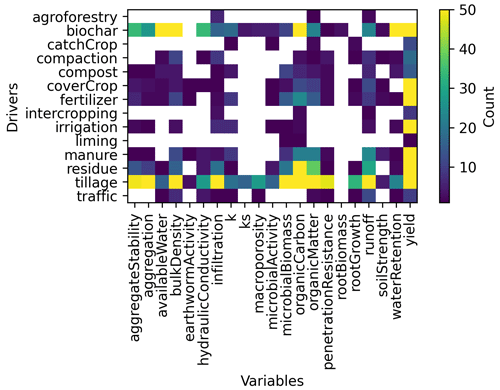
Figure 7 shows the number of relationships from abstracts extracted according to the pair driver/variable identified within them. Relationships including “biochar” or “tillage” as drivers were the most frequent, while “yield” was the variable most commonly found. Note as well that for some combinations of drivers/variables, no statements were available. This helped to identify knowledge gaps within our corpus. For instance, the effect of liming on aggregates and infiltration properties was not studied in our corpus. Similarly the effect of irrigation on soil organic carbon was also not present in the corpus.
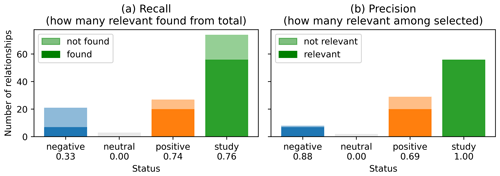
Figure 8Recall (a) and precision (b) of classified relationships extracted from abstracts. Dark color represents the proportion of relationships correctly classified, while the faded color represents relationships not found or not correctly classified. The recall and precision metric for each category is given on the x axis. Results were obtained for the Meta corpus.
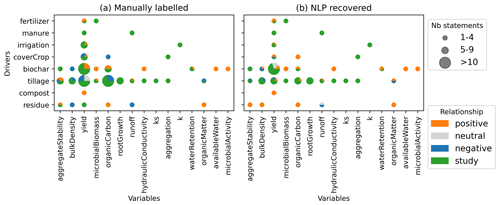
Figure 9Relationships between drivers and variables as (a) manually labeled and (b) recovered by NLP for the Meta corpus.
However, one important limitation of the approach is that the algorithm can only find the keywords it was told to look for. For instance, no statement including social drivers were found by our method as we did not add keywords associated with social drivers. Nevertheless, we acknowledge the importance of social drivers to estimate the acceptability of management practices (EJP SOIL – CLIMASOMA, 2022, chapter 3), and they would gain to be included in future workflows. Another limitation is the fact that the algorithm is limited to what is written in the text. For instance, in Fig. 7, the tokens “k”, “Ks” and “hydraulic conductivity”, all associated with hydraulic conductivity, are all extracted by the NLP algorithm as they appear in this form in the abstracts. The use of synonyms can help associate tokens with similar meaning.
Figure 8 shows the recall and the precision of the extracted relationships according to their labeled status. For each category (negative, neutral, positive or study), the dark color represents the proportion of relationships correctly identified by the NLP algorithm. The faded color represents the relationships wrongly classified by the NLP or not found at all. Overall, most identified relationships belong to the “study” class. Note as well the larger number of “positive” relationships compared to “negative”, which may be a manifestation of some bias in reporting positive results or at least writing them as positive relationships. The precision of the NLP algorithm was high for “negative” (precision = 0.88) or “study” (precision = 1.00) classes. In terms of recall, the highest score is achieved for both “positive” and “study” categories.
Based on manually labeled relationships and the ones recovered from NLP, Fig. 9a offers a detailed comparison according to the number of statements recovered (size of the bubble) and their correlations (colors). Such a figure has the potential to be used to get a first overview of the relationships present in a large corpus of studies (e.g., for evidence synthesis). It is also comparable to figures presented in the report EJP SOIL – CLIMASOMA (2022, chapter 1), which presents a similar layout with the results from the selected meta-analysis. Note that not all statements have the same relationships for specific driver–variable pairs (not all studies have the same conclusions), which causes the bubbles in Fig. 9 to contain multiple colors (e.g., biochar/yield, tillage/runoff). According to the relationship extraction, compost addition was positively correlated to yield, residues were positively associated with lower bulk density and lower run-off, and biochar was negatively correlated to bulk density and positively correlated to microbial biomass. Most of these relationships correspond well to what is reported in meta-analysis (EJP SOIL – CLIMASOMA, 2022, chapter 1). As demonstrated already in Fig. 8, the NLP did not recover all relationships perfectly (low recall for negative relationships) and can sometimes be completely wrong (e.g., residue/bulk density). But in two-thirds of all cases (66 %), the relationships were classified correctly.
Relationship extraction based on abstracts provides a quick overview of the conclusions from a given set of documents (Fig. 9). However, the classification of the extracted relationships remains a challenging task, and a lot of statements just mention that the driver–variable pair has been studied but not the outcome of it. That is one of the limitations of the approach as not all information is contained in the abstract. Applying this technique on the conclusion part of a paper could help complement the relationships found.
In addition, to confirm that the relationships extracted are well classified, one has to manually label a given proportion of the statements found and then compare the labels with the NLP finding and iteratively improve the NLP algorithm. This procedure is tedious, but needed, as general relationships algorithms (often trained on newspaper articles or Wikipedia) failed to extract meaningful relationships from field-specific scientific publications. This is in agreement with the conclusions by Furey et al. (2019). However, despite our efforts, the complexity of certain sentences (long sentences with comparison and relative clauses) was too high for our algorithm to reliably detect the relationships between a driver and a variable.
With the growing body of environmental scientific literature, NLP techniques can help support the needed evidence synthesis. We explored practical applications of NLP to classify documents into topics, identify knowledge gaps, build databases using regular expression and extract the main conclusion of the abstract through relationship extraction. While NLP techniques cannot replace human intervention, their automatic nature enables us to quickly process a large corpus of scientific publications. When compiling an evidence synthesis, one can start by querying online search engines with specific query strings. Sets of documents can then be analyzed using topic modeling, and newer publications can be classified into the found topics. This approach enables us to identify possible knowledge gaps or topics less studied. A second step would be to extract a set of specific contextual information. In this work, we demonstrated the usefulness of simple regular expressions for these tasks. Instead of manually entering data into a database form, the algorithm could prefill the form for the user to verify. The database produced can later be used for more quantitative analysis such as meta-analysis or machine learning techniques. Finally, a third step would be to extract the main conclusion of the publications. While natural language understanding is a fast-growing field, the relationship extraction algorithm developed in this work was already able to extract and classify pairs of practices (drivers) and variables (soil and site properties). While their classification remains challenging and field specific given the complexity of human language, this approach already provides a good overview of the main conclusions drawn from a corpus of documents.
Overall, the NLP techniques presented in this work have practical potential to support high-throughput semi-automated evidence synthesis that can be continuously updated as new publications become available. Given sufficient training data, the use of more advanced methods that convert sentences to numerical vectors by the use of transformer networks (e.g., BERT; Koroteev, 2021), coupled with deep learning algorithms, presents new and exciting possibilities for language understanding.
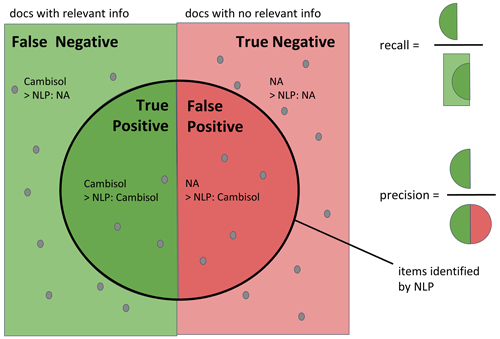
Figure A1Schematic representation of precision and recall. Recall aims to assess how much relevant information was selected out of all the data available in the corpus, while precision aims to assess how much relevant information was in the selection.
All processing data and figures presented in this article are available in the form of Jupyter Notebooks: https://doi.org/10.5281/zenodo.7687794. The authors of this paper are the authors of the repository. Due to copyright restriction, the papers are not provided, but a list of references used is available on the GitHub repository.
Conceptualization: GB. Data curation: GB, LA and JK. Formal analysis: GB. Project administration: JK and SG. Writing original draft: GB. Writing review and editing: GB, JK and SG.
The contact author has declared that none of the authors has any competing interests.
Publisher’s note: Copernicus Publications remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This work was developed inside the CLIMASOMA project from the EJP SOIL consortium. EJP SOIL has received funding from the European Union's Horizon 2020 research and innovation program (grant agreement no. 862695) and was co-funded by the involved partners: ILVO, Agroscope, SLU, WUR and CREA.
This paper was edited by Olivier Evrard and reviewed by two anonymous referees.
Angeli, G., Johnson Premkumar, M. J., and Manning, C. D.: Leveraging Linguistic Structure For Open Domain Information Extraction, in: Proceedings of the 53rd Annual Meeting of the Association for Computational Linguistics and the 7th International Joint Conference on Natural Language Processing (Volume 1: Long Papers), Proceedings of the 53rd Annual Meeting of the Association for Computational Linguistics and the 7th International Joint Conference on Natural Language Processing (Volume 1: Long Papers), Beijing, China, 344–354, https://doi.org/10.3115/v1/P15-1034, 2015.
Caracciolo, C., Stellato, A., Morshed, A., Johannsen, G., Rajbhandari, S., Jaques, Y., and Keizer, J.: The AGROVOC linked dataset, AGROVOC, 4, 341–348, 2013.
EJP SOIL – CLIMASOMA: CLIMASOMA – Final report Climate change adaptation through soil and crop management: Synthesis and ways forward, https://climasoma.curve.space/report (last access: 1 March 2023), 2022.
Furey, J., Davis, A., and Seiter-Moser, J.: Natural language indexing for pedoinformatics, Geoderma, 334, 49–54, https://doi.org/10.1016/j.geoderma.2018.07.050, 2019.
Haddaway, N. R., Callaghan, M. W., Collins, A. M., Lamb, W. F., Minx, J. C., Thomas, J., and John, D.: On the use of computer-assistance to facilitate systematic mapping, Campbell Systematic Reviews, 16, e1129, https://doi.org/10.1002/cl2.1129, 2020.
Hirschberg, J. and Manning, C. D.: Advances in natural language processing, Science, 349, 261–266, https://doi.org/10.1126/science.aaa8685, 2015.
Honnibal, M. and Montani, I.: spaCy 2: Natural language understanding with bloom embeddings, convolutional neural networks and incremental parsing, To Appear, 7, 411–420, 2017.
Jarvis, N., Koestel, J., Messing, I., Moeys, J., and Lindahl, A.: Influence of soil, land use and climatic factors on the hydraulic conductivity of soil, Hydrol. Earth Syst. Sci., 17, 5185–5195, https://doi.org/10.5194/hess-17-5185-2013, 2013.
Koroteev, M. V.: BERT: A Review of Applications in Natural Language Processing and Understanding (arXiv:2103.11943), arXiv, https://doi.org/10.48550/arXiv.2103.11943, 2021.
Lee, J., Yoon, W., Kim, S., Kim, D., Kim, S., So, C. H., and Kang, J.: BioBERT: a pre-trained biomedical language representation model for biomedical text mining, Bioinformatics, 36, 1234–1240, https://doi.org/10.1093/bioinformatics/btz682, 2020.
Lin, J.: Divergence measures based on the Shannon entropy, IEEE T. Inform. Theory, 37, 145–151, https://doi.org/10.1109/18.61115, 1991.
Loper, E. and Bird, S.: NLTK: The Natural Language Toolkit (arXiv:cs/0205028), arXiv, https://doi.org/10.48550/arXiv.cs/0205028, 2002.
Manning, C., Surdeanu, M., Bauer, J., Finkel, J., Bethard, S., and McClosky, D.: The Stanford CoreNLP Natural Language Processing Toolkit, in: Proceedings of 52nd Annual Meeting of the Association for Computational Linguistics: System Demonstrations, Proceedings of 52nd Annual Meeting of the Association for Computational Linguistics: System Demonstrations, Baltimore, Maryland, 55–60, https://doi.org/10.3115/v1/P14-5010, 2014.
Nadkarni, P. M., Ohno-Machado, L., and Chapman, W. W.: Natural language processing: an introduction, J. Am. Med. Inform. Assoc., 18, 544–551, https://doi.org/10.1136/amiajnl-2011-000464, 2011.
Nasar, Z., Jaffry, S. W., and Malik, M. K.: Information extraction from scientific articles: a survey, Scientometrics, 117, 1931–1990, https://doi.org/10.1007/s11192-018-2921-5, 2018.
Niklaus, C., Cetto, M., Freitas, A., and Handschuh, S.: A Survey on Open Information Extraction (arXiv:1806.05599), arXiv, https://doi.org/10.48550/arXiv.1806.05599, 2018.
Padarian, J. and Fuentes, I.: Word embeddings for application in geosciences: development, evaluation, and examples of soil-related concepts, SOIL, 5, 177–187, https://doi.org/10.5194/soil-5-177-2019, 2019.
Padarian, J., Minasny, B., and McBratney, A. B.: Machine learning and soil sciences: a review aided by machine learning tools, SOIL, 6, 35–52, https://doi.org/10.5194/soil-6-35-2020, 2020.
Ramakrishnan, C., Patnia, A., Hovy, E., and Burns, G. A.: Layout-aware text extraction from full-text PDF of scientific articles, Source Code Biol. Med., 7, 7, https://doi.org/10.1186/1751-0473-7-7, 2012.
Rastan, R., Paik, H.-Y., and Shepherd, J.: TEXUS: A unified framework for extracting and understanding tables in PDF documents, Info. Proc. Manage., 56, 895–918, https://doi.org/10.1016/j.ipm.2019.01.008, 2019.
Röder, M., Both, A., and Hinneburg, A.: Exploring the Space of Topic Coherence Measures, in: Proceedings of the Eighth ACM International Conference on Web Search and Data Mining, WSDM 2015: Eighth ACM International Conference on Web Search and Data Mining, Shanghai China, 399–408, https://doi.org/10.1145/2684822.2685324, 2015.
Sievert, C. and Shirley, K.: LDAvis: A method for visualizing and interpreting topics, in: Proceedings of the Workshop on Interactive Language Learning, Visualization, and Interfaces, Proceedings of the Workshop on Interactive Language Learning, Visualization, and Interfaces, Baltimore, Maryland, USA, 63–70, https://doi.org/10.3115/v1/W14-3110, 2014.
Tao, C., Filannino, M., and Uzuner, Ö.: Prescription Extraction Using CRFs and Word Embeddings, J. Biomed. Inform., 72, 60–66, https://doi.org/10.1016/j.jbi.2017.07.002, 2017.
Wang, Y., Wang, L., Rastegar-Mojarad, M., Moon, S., Shen, F., Afzal, N., Liu, S., Zeng, Y., Mehrabi, S., Sohn, S., and Liu, H.: Clinical information extraction applications: A literature review, J. Biomed. Inform., 77, 34–49, https://doi.org/10.1016/j.jbi.2017.11.011, 2017.