the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
Ectomycorrhizal fungal network complexity determines soil multi-enzymatic activity
Jorge Prieto-Rubio
José L. Garrido
Julio M. Alcántara
Concepción Azcón-Aguilar
Ana Rincón
Álvaro López-García
Soil functioning is intrinsically related to the structure of associated biological communities. This link is barely understood in the multispecies context of soil microbial communities, which often requires complex analytical approaches to discern structural and functional roles of microbial taxa inhabiting the soil. To investigate these ecological properties, we characterized the assembly and soil functioning contribution of ectomycorrhizal (ECM) fungal communities through co-occurrence network analysis. Co-occurrence networks were inferred from ECM root tips of Cistus albidus, Quercus faginea and Q. ilex on a regional scale, in Mediterranean mixed forests. Soil enzymatic activities related to carbon and nutrient cycling were also measured, and soil functionality outcomes related to ECM fungal network structure were evaluated on the community to taxon levels. Network complexity relied on habitat characteristics and seasonality, and it was linked to different dominant ECM fungal lineages across habitats. Soil enzymatic activities were habitat-dependent, driven by host plant identity and fungi with reduced structuring roles in the co-occurrence network (mainly within Thelephorales, Sebacinales and Pezizales). ECM fungal co-occurrence network structure and functioning were highly context-dependent, pointing to divergent regional fungal species pools according to their niche preferences. As increased network complexity was not related to greater soil functionality, functional redundancy might be operating in Mediterranean forest soils. The revealed differentiation between structural and functional roles of ECM fungi adds new insights into the understanding of soil fungal community assembly and its functionality in ecosystems.
- Article
(1213 KB) - Full-text XML
-
Supplement
(994 KB) - BibTeX
- EndNote
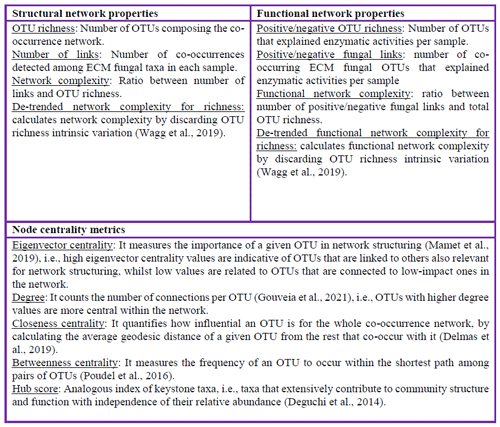
The structure of biological communities is a key element for predicting the potential role of biodiversity in ecosystem functioning (Wagg et al., 2021). The ways through which species are assembled, e.g., via environmental filters, disturbances or even demographic stochasticity, can lead their functions within an ecosystem (Mazel et al., 2018; Wang et al., 2022). Several studies have attempted to link the structure of biological communities to their functionality, such as those relating taxonomic or phylogenetic diversity to functional traits of species or to multifunctionality indices (Delgado-Baquerizo et al., 2016; Le Bagousse-Pinguet et al., 2019; Liu et al., 2022). Some of the most widespread tools to associate community metrics with ecosystem functions are based on correlations across variables or regression models that allow one to infer ecosystem function responses through community structure predictors, e.g., species richness or phylogenetic structure (Pérez-Izquierdo et al., 2017; Bastida et al., 2019; Krapu and Borsuk, 2020). However, most community structure metrics do not consider how the occurrences of species, and even their abundance, are affected by the presence or absence of other species and to what extent this species co-occurrence might explain ecosystem functions (Wagg et al., 2019). Within this context, network analysis has emerged as a promising analytical approach to studying multispecies co-occurrence and, hence, to evaluating assembly rules, niche differentiation and expected functionality of integrated species assemblages (Mandakovic et al., 2018; Zhan et al., 2021; Goberna and Verdú, 2022).
Co-occurrence network analysis is particularly useful on data obtained from next-generation sequencing at the level of operational taxonomic units (OTUs), species hypothesis (SH) or amplicon sequence variants (ASVs) as species equivalents (Barberán et al., 2012; Barroso-Bergadà et al., 2020). However, this approach is affected by biases, e.g., due to the sampling differences or intrinsic biotic and abiotic agents triggering indirect interactions that may result in spurious associations among species (Gloor et al., 2017; Barner et al., 2018; Barroso-Bergadà et al., 2020). To accurately infer species associations and overcome these constraints, several tools have been developed by applying distribution modeling or integrating correlation-based algorithms, such as the SParse InversE Covariance Estimation for Ecological Association Inference (SPIEC-EASI) (Kurtz et al., 2015; Morueta-Holme et al., 2016; Watts et al., 2019). These analytical frameworks have proven useful for characterizing environmental microbial communities, usually involving hyperdiverse communities with key implications for ecosystems (Pauvert et al., 2019; Wagg et al., 2019; Barroso-Bergadà et al., 2020; Luo et al., 2022).
Among soil microbial communities, ectomycorrhizal (ECM) fungi exhibit high taxonomic and functional diversity in forest ecosystems (Nguyen et al., 2020). ECM fungi mainly form symbiotic interactions in the roots of tree and shrub lineages and are known to promote positive plant–soil feedbacks (Kadowaki et al., 2018; Bahram et al., 2020; Liang et al., 2020; Segnitz et al., 2020). In the ECM symbiosis, the plant delivers photosynthetic carbon to the fungus in exchange for nutrients that the fungus obtains through the external mycelium spreading in the soil (Smith and Read, 2008). ECM fungi are capable of scavenging nutrients from soil organic matter by producing exoenzymes (Pritsch and Garbaye, 2011) which vary across ECM fungal lineages, promoting contrasting nutrient cycling and soil functional outcomes (Miyauchi et al., 2020; Lebreton et al., 2021; Prieto-Rubio et al., 2023). In addition, the C-enriched environment generated by the ECM symbiosis, i.e., mycorrhizosphere or hyphosphere (sensu van der Heijden et al., 2015), may harbor microorganisms (e.g., bacteria and saprotrophic fungi) with enzymatic capabilities to decompose organic matter, including ECM symbiotic structures and mycelia (Azcón-Aguilar and Barea, 2015; Cheeke et al., 2017; Lebreton et al., 2021; Nguyen, 2023). Hence, as the ECM environment seems key for carbon and nutrient cycling, understanding the structure of their co-occurrence networks might bring new insights into how ECM fungal species are assembled within the communities and how they contribute to explaining forest soil functioning (Barroso-Bergadà et al., 2020; Wagg et al., 2021, Martin and van der Heijden, 2024).
On the basis that the structure of ECM fungal communities is driven by habitat conditions and host plant community composition (Glassman et al., 2017; Pérez-Izquierdo et al., 2018; Wu et al., 2018), we firstly hypothesized that ECM fungal co-occurrence network structure would be dependent on host plant identity and spatial–temporal factors (habitat and seasonality). The key role of ECM fungal communities in soil functioning and soil biodiversity maintenance (Courty et al., 2010; Miyauchi et al., 2020) was the origin of our second hypothesis: the richer and/or more complex the ECM fungal co-occurrence network, the greater the soil functionality (i.e., carbon and nutrient cycling). This would be based on the associated fungal taxa with contrasting profiles of enzymatic activity leading to functional overdispersion as a result of niche segregation (Götzenberger et al., 2012; Wagg et al., 2019).
To test these hypotheses, we characterized the ECM fungal co-occurrence networks of Mediterranean mixed forests of southern Spain, at individual host plant levels from three representative plant species, and measured different potential soil extracellular enzymatic activities as a proxy of carbon and nutrient mobilization. We aimed to (i) infer the ECM fungal co-occurrence networks and study their structure and complexity across contrasting scales (forest, plot, host plant individual and ECM fungal taxon), (ii) determine the impact of ECM fungal co-occurrence network complexity on soil functionality and (iii) estimate the relationship between co-occurring ECM fungal taxa and the associated enzymatic activities in the soil.
2.1 Study sites
This study was conducted in Mediterranean mixed forests located in the natural parks of Sierras de Cazorla, Segura y Las Villas (38.29° N, −2.57° W, referred to hereafter as Segura) and Monte la Sierra (37.64°N, −3.73° W, referred to hereafter as Jaén), in southern Spain. These protected areas have been largely monitored for plant–plant and plant–microbe interactions to better understand topics concerning Mediterranean forest community dynamics (from Siles et al., 2008, to Garrido et al., 2023, and Pajares-Murgó et al., 2023). With average altitudes of 1300 and 1000 m in Segura and Jaén, respectively, the climate in both regions is continental Mediterranean, showing annual mean temperatures of 13–15 °C and rainfall of 550–600 mm, with cool wet winters and summer droughts. Soils are characterized by calcareous limestone and low nutrient retention. The canopy layer mixes Quercus ilex L. and Q. faginea Lam. with Pinus halepensis Mill. in Jaén and with Q. pyrenaica Willd., Pinus nigra subsp. salzmanii J. F. Arnold and Pinus pinaster Ait. in Segura. At both sites, the understory includes Juniperus spp. and tree and shrub species of Cistaceae, Oleaceae, Rosaceae and Lamiaceae, among others (Pulgar et al., 2017; Alcántara et al., 2018). To compare ECM fungal community network functionality outcomes across the study sites, we selected those ECM plants that were common at both study sites (i.e., Cistus albidus L., Q. faginea and Q. ilex).
2.2 Experimental design and sampling
Four 50×50 m plots were selected per forest, and samplings were carried out in fall (2016) and spring (2017) (Alcántara et al., 2018). A total of 92 adult plant individuals were sampled, 32 for Q. ilex, 31 for Q. faginea and 29 for C. albidus (47 in Jaén and 45 in Segura, and 48 in fall and 44 in spring), with at least 10 m distance between the given plots. After removing the litter layer, we collected three subsamples ( cm holes) with secondary roots tracked from the target plant and surrounding soil and further combined into a single composite sample for each individual (Prieto-Rubio et al., 2022). Once in the lab, roots were separated from the surrounding soil and washed with tap water over 2 and 0.5 mm sieves, and all ECM root tips were collected under a Carl Zeiss Stemi 2000 stereomicroscope (Rincón et al., 2014). Before physical–chemical analyses, the collected soil in each sample was homogenized, air-dried and 2 mm sieved. The soil pH was measured in solutions (1:5, w:v in H2O), the gravimetric moisture (GM) was calculated as the difference in soil weight before and after drying the samples at 105 °C for 48 h, and the soil organic matter (SOM) was determined by measuring weight loss on ignition at 400 °C for 4 h, following the methodology in Walkley and Black (1934) (see the details of the soil physical–chemical analyses in Prieto-Rubio et al., 2022).
2.3 ECM fungal community and soil multi-enzymatic activity
To frame the ECM fungal community in co-occurrence networks and relate it to potential soil enzymatic activities, we used the raw data (ITS1 sequences by Illumina MiSeq) of ECM fungal communities present at root tips and the enzymatic activities of soils collected in the rhizosphere of C. albidus, Q. ilex and Q. faginea, both at Jaén and Segura. The taxonomy of fungal amplicon sequence variants was assigned using the UNITE database v7.2 (Cole et al., 2014; Abarenkov et al., 2010) and clustered at 97% similarity into OTUs. After filtering the obtained fungal OTUs using the FUNGuild database v1.1 (Nguyen et al., 2016), the final output resulted in 449 OTUs with 6 582 941 reads associated with the ECM lifestyle. Two independent ECM abundance matrices, one per study forest, Jaén or Segura, were used for subsequent network analyses. Further details on molecular and bioinformatics analyses are provided as Supplement S1.
Nine potential extracellular enzymatic activities related to C, N and P cycling in forest soils were measured: β-glucosidase, cellobiohydrolase, β-xylosidase, β-glucuronidase and laccase related to C cycling; acid and alkaline phosphatase activities promoting P cycling; and chitinase and leucine-aminopeptidase activities as proxies of N cycling. Laccase was determined through a photometric assay and the rest through fluorogenic assays using a Victor microplate reader (Perkin-Elmer Life Sciences, Massachusetts, USA) (see details in Supplement S1). A multi-enzymatic activity index (MAI) was calculated from the enzymatic activities' dataset with a principal component analysis (PCA)-based procedure described in Meyer et al. (2018). Briefly, each enzymatic activity was standardized into z scores with the decostand function in the vegan R package (Oksanen et al., 2022). Then, a PCA was run with the prcomp function over the z-score dataset in the stats R package, and PC axes were multiplied by the proportion of variation that each captured (i.e., weighted PC scores). To ensure that higher scores corresponded to higher enzymatic activities, the signs of the scores on each PC axis were inspected and the whole axis was multiplied by −1 when necessary (reoriented PC scores, Meyer et al., 2018; Wagg et al., 2019). The MAI of each sample was calculated by summing its reoriented weighted PC scores.
2.4 Co-occurrence networks in ECM fungal communities
In our study, we followed the recommendations of Goberna and Verdú (2022) concerning the use of a co-occurrence concept for soil microbial communities, instead of other studies that assume direct association and/or interaction between the organisms. To infer the co-occurrence of ECM fungi within the communities, we adjusted the methodology as described by Wagg et al. (2019), which allows associations among pairs of OTUs to be sought and framed under the conditional independence concept (Kurtz et al., 2015). The OTU abundance matrix was rescaled by the proportion of minimum sequencing depth (39 221 sequences in Jaén and 4076 in Segura; see Prieto-Rubio et al., 2022, for more details on sequencing yielding), and OTUs that appeared <1 % in relative abundance were discarded. We obtained two curated datasets with 47 (Jaén) and 66 (Segura) ECM fungal OTUs that were used to infer the co-occurrence matrix of each ECM fungal community by applying the SPIEC-EASI algorithm with the spiec.easi function in the SpiecEasi R package (Kurtz et al., 2015). This is a robust correlation-based method for ecological network inference that differs from others in discarding spurious associations due to the lack of independence, i.e., correlations among OTUs that might be indirectly affected by other OTUs occurring in the network (Kurtz et al., 2015). The networks were constructed with the Meinshausen and Bühlmann (2006) neighborhood selection method. The minimum lambda ratio (i.e., a scalar tuning parameter in high-dimensional data) was 0.01, and 50 values of lambda for every 100 cross-validation permutations were fitted to evaluate the most parsimonious networks by using the StARS model selection criterion (Liu et al., 2010).
From the global adjacency co-occurrence matrices of each forest (spiec.easi objects), we extracted the presence–absence adjacency co-occurrence matrix per sample by pruning the global matrix to only include the OTUs in each sample. At the host plant individual level, we determined the OTU richness, the number of links (i.e., the number of co-occurrences detected among ECM fungal taxa in each sample) and the ratio number of links and OTU richness that were used as proxies of network complexity in each sample (Box 1). We then built weighted networks for the global adjacency matrices with the graph.adjacency function in the igraph R package (Csardi and Nepusz, 2006), with edge weight representing the co-occurrence frequency of ECM fungal OTUs. The adjacency matrices were used to graphically represent the networks with the Gephi software v.0.9.2 (Bastian et al., 2009). Both the Fruchterman–Reingold distribution and ForceAtlas2 attraction–repulsion algorithms were used in the simulations (Fruchterman and Reingold, 1991; Jacomy et al., 2014). We obtained the network graph and calculated different network centrality metrics at the ECM fungal OTU level (degree, eigenvector, closeness and betweenness centralities) and the hub score as analogous to a keystone taxa index (see Box 1). These metrics were used to measure the contribution of each OTU in structuring the co-occurrence network at site level. Network centrality metrics were performed through several algorithms in Gephi, all of them scaled from 0–1 (Box 1). Further, we determined the modules (i.e., sub-communities integrating the ECM fungal co-occurrence network per site) to which the OTUs belonged by using the algorithm described in Blondel et al. (2008) and implemented in Gephi.
2.5 Statistical analyses
2.5.1 ECM fungal network structure and habitat characteristics (hypothesis 1)
The response of network complexity to the factors studied was evaluated for each forest (Jaén and Segura) separately. Seasonality was fitted as a fixed factor and host plant identity nested in plots as a random factor by means of linear mixed-effects models (LMMs), using the lme4 R package (Bates et al., 2015). The significance of explanatory factors was tested with the Anova and rand functions from the lmerTest and car R packages, respectively. The variance explained of both fixed and random factors was evaluated with the r.squaredGLMM function in the MuMIN R package. In addition, the relationships of soil properties (log-transformed pH, SOM and GM) and network complexity parameters were assessed through Spearman correlation tests with Bonferroni adjustment using the psych and ggcorrplot R packages. The relationships between OTU-level network metrics were also evaluated by following the same correlation and adjustment tests.
To determine the importance of OTU abundance for network structuring (Carvalhais and Dennis, 2021), its effect on network centrality metrics (Box 1) was analyzed by means of permutational ANOVA (PERMANOVA) (adonis function, vegan R package) using Bray–Curtis dissimilarity with 999 permutations. Besides, to evaluate whether some modules were more central than others within the ECM fungal networks, we tested centrality metrics of OTUs with the corresponding module ID using PERMANOVA.
2.5.2 ECM fungal networks and soil multi-enzymatic activity (hypothesis 2)
To detect whether the structure of ECM fungal co-occurrence networks explained soil functionality (i.e., carbon and nutrient cycling), the response of the MAI was evaluated with LMMs by considering OTU richness, the number of links or network complexity (i.e., metrics determined at the host plant individual level) and seasonality as fixed factors. The response of detrended network complexity, which discards the intrinsic variation derived from OTU richness, was also evaluated with the same model and factors. The plant species nested in the plot was considered a random factor in all LMMs.
The ECM fungal OTUs potentially affecting each soil enzymatic activity were detected by the elastic net (ENET) regularization method (Zou and Hastie, 2005; Lassalle et al., 2019; Wagg et al., 2019). Briefly, the ENET regularization combines LASSO (least absolute shrinkage and selection operator) and Ridge penalty-based regression modeling, allowing us to avoid overfitting and the correlation effects between predictors (i.e., the OTUs embedded in the co-occurrence networks) and to minimize the influence of those that explain the variations in the response variables to a lesser extent (i.e., each of the soil enzymatic activities) (Zou and Hastie, 2005). As a previous step before model training, the abundance matrix of ECM fungal OTUs by host plants was standardized. Significant effects were determined by the standardized effect size | SES| >1.96 (i.e., the comparison of the observed effect of the given OTU with the simulated ones – 999 iterations – divided by the standard deviation of the simulated ones). From the ENET results, we recorded all the OTUs predicting enzymatic activities (i.e., those that contributed to predicting a given enzymatic activity) and determined the OTU richness that explained enzymatic activities per host plant individual. Further, we could determine the number of co-occurring ECM fungal OTUs that supported a common enzymatic activity (i.e., positive or negative fungal links) and the functional network complexity by dividing positive or negative fungal links by the total OTU richness (Box 1). In addition, as fungal network properties can be affected by the species or OTU number, especially when this decreases (Berry and Widder, 2014), the detrended network parameters by ECM fungal OTU richness allowed us to evaluate how the co-occurrence network structure yielded multi-enzymatic activity responses by discarding the variance intrinsically derived from the OTU richness. All functional network parameters recorded in each study forest (i.e., positive or negative OTU richness, positive or negative fungal links, and positive or negative complexity) and their detrended values for the total OTU richness were used in linear regression models (LMs) to explore their effects on MAI responses by fitting a separate LM for each functional parameter with the lm function in the stats R package.
3.1 Structure of ECM fungal co-occurrence networks
Contrasting results in network parameters were observed between the study forests (Table 1). In Jaén, seasonality significantly affected ECM fungal network complexity, i.e., lower in spring than in fall (Table 1, Fig. S1 in the Supplement), while in Segura, this effect was detected in the OTU richness and the number of fungal links, in both cases higher in spring than in fall. Host plant species identity affected the detrended network complexity at the genus level in Segura (Table 1) (Cistus and Quercus spp. respective scores of 0.16 and 0.22, , p=0.05).
Table 1Effects of spatial–temporal factors and host plant species on overall ectomycorrhizal fungal network properties inferred from spiec.easi.
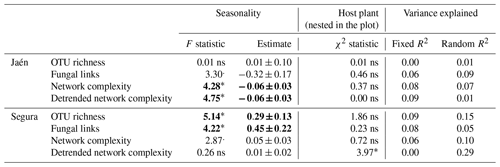
Linear mixed models were separately performed for each study forest: Jaén and Segura. Network properties (see Box 1) were log-transformed; seasonality was fitted as a fixed factor and host plant species nested in the plot as a random factor. The effects of fixed and random factors were determined by the F and χ2 statistics, respectively, and the variance explained by both factor pools (R2). Significance level: “” p<0.001, “” p<0.01, “*” p<0.05, “.” p<0.10 and “ns” – nonsignificant (significant effects in bold).
When the forest was analyzed as a fixed factor, the ECM fungal network complexity was revealed to be significantly higher in Segura than in Jaén (Fig. S2). In fact, the network complexity and soil properties of each site were differentially correlated (Fig. S3). In general, soil pH was positively related to network parameters in Jaén and negatively in Segura, while higher SOM or GM values were associated with lower network complexity in both Jaén and Segura. The number of modules of the inferred co-occurrence networks varied between forests (Fig. 1, Table S1 in the Supplement). In Jaén, ECM fungal OTUs were distributed across 33 modules, 27 % of them composed of more than one OTU, and none of them was more central than the others (PERMANOVA, , p=0.67, R2=0.01). By contrast, ECM fungal OTUs were distributed across 27 modules in Segura, 44 % of them with two or more OTUs, and some (e.g., modules 2, 18 or 22) were more central than the rest (PERMANOVA, , p=0.02, R2=0.08) (Fig. 1, Table S1).
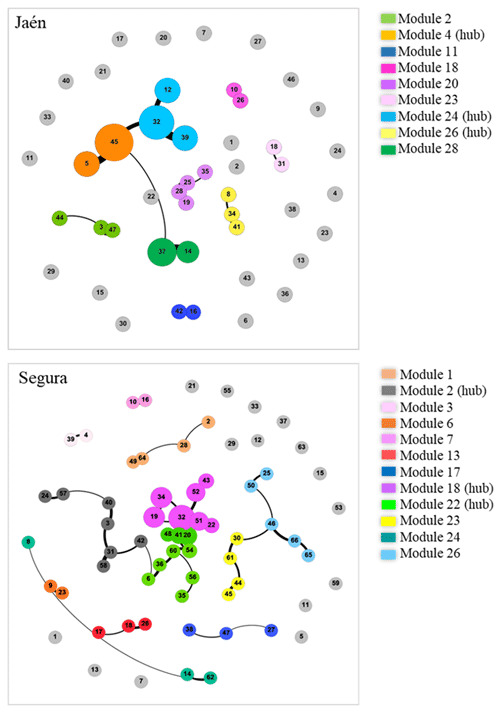
Figure 1Co-occurrence networks of ectomycorrhizal (ECM) fungal communities in each study forest: Jaén and Segura. Graphs are inferred through the Gephi software by using the Fruchterman–Reingold and ForceAtlas algorithms. In each site network, each node number represents an ECM fungal OTU (see Table S1 for the taxonomic assignment and network metrics for each forest), and the node size is related to the hub score index (i.e., a proxy of keystone taxa). Co-occurring OTUs are connected by edges whose width indicates the co-occurrence strength (co-exclusions were not detected among ECM fungal taxa across the study sites). Within each forest ECM fungal community, a shared node color indicates a separate module (legends) inferred by Gephi (Table S1). Nodes in light-grey color indicate non-co-occurring OTUs in each forest network.
Negative correlations between the average abundance of ECM fungal OTUs and network centrality metrics (degree, closeness, eigenvector and betweenness centralities) were observed at both sites (Fig. 2) (PERMANOVA , p=0.02, R2=0.09 in Jaén; , p<0.001, R2=0.11 in Segura). Correlations between centrality metrics were all positive and overall greater in Jaén than in Segura (Fig. 2). In addition, hub scores (i.e., a proxy of keystone taxa; Box 1) were observed in specific Gephi modules, particularly in modules 4, 24 and 28 in Jaén and in modules 2, 18 and 22 in Segura (Fig. 1, Table S1). In Jaén, a total of seven hub OTUs were detected, most belonging to Thelephoraceae (six OTUs) (e.g., Tomentella with four) and one to Russulaceae (Table S1). However, in Segura, a widespread taxonomic diversity of hub OTUs (23 in total) across different fungal families and genera, e.g., Inocybe (7), Tomentella (5), Sebacina (4), Tuber (1), Tylospora (1), Amphinema (1), Pustularia (1), Humaria (1) and Hydnum (1), was observed (Table S1).
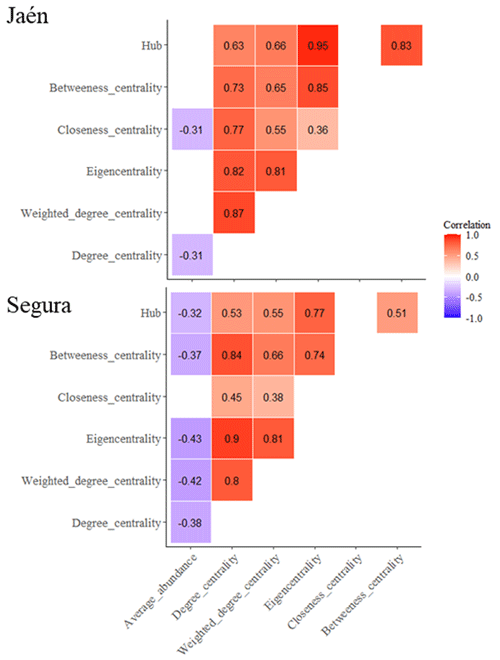
Figure 2Correlations between average ECM fungal OTU abundance and network parameters (Box 1). Network metrics calculated for each ECM fungal OTU are inferred through the Gephi software. Correlation tests are calculated with the Spearman method using the Bonferroni adjustment for the dataset of each study forest: Jaén and Segura. Significant correlations (p<0.05) are plotted per site, indicating positive (red) or negative (blue) correlations.
3.2 Functionality of ectomycorrhizal co-occurrence networks
When functional ECM fungal co-occurrence network parameters were tested as explanatory variables of the multi-enzymatic activity MAI, a number of significant effects – positive OTU richness, detrended positive and negative OTU richness, functional network complexity – were observed, but only in Jaén (Table 2). In Segura, only positive OTUs were recorded, although they did not significantly affect the soil MAI (Table 2); no effect of the general ECM fungal network parameters (i.e., fungal links and network complexity) on the MAI was detected (Table S2).
Table 2Effects of functional network parameters inferred from elastic net (ENET) regularization models on the soil multi-enzymatic activity index (MAI) per study forest: Jaén and Segura.
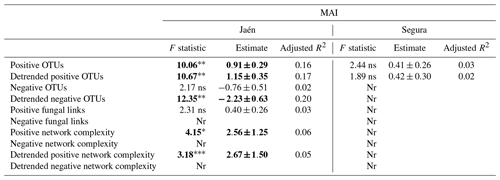
Notes: each network metric was fitted as a predictor of the soil MAI in linear models when recorded from the given forest. Detrended functional network metrics were also fitted as predictors. Significant values are highlighted in bold: “” p<0.01, “*” p<0.05, “” p<0.10 and “ns” >0.10. Nr: not recorded.
The number of OTUs that explained at least one soil enzymatic activity differed greatly between the study forests, i.e., 76.6 % and 9.1 % of their respective total OTUs in Jaén and Segura, respectively (Fig. 3, Table S3). In both forests, ECM taxa that explained soil enzymatic activities showed low centrality scores in the co-occurrence networks (Tables S1, S3). Additionally, keystone taxa (i.e., those showing hub scoring, Fig. 1) also recorded low effects on the enzymatic activities (Fig. 3). In Jaén, 36 OTUs positively or negatively predicted soil enzymatic activities, particularly those related to C cycling (Fig. 3a, Table S3a). Most of these OTUs belonged to Thelephorales (23.40 %), Sebacinales (21.28 %) and Pezizales (12.77 %) and in many cases affected more than one enzymatic activity, as was also the case with the unique OTU belonging to Boletales (Fig. 3, Table S3a). Fungi within the Pezizales order showed a predominantly positive effect, particularly on enzymatic activities related to C (β-glucosidase and laccase) and P (alkaline phosphatase) cycling (Fig. 3, Table S3a).
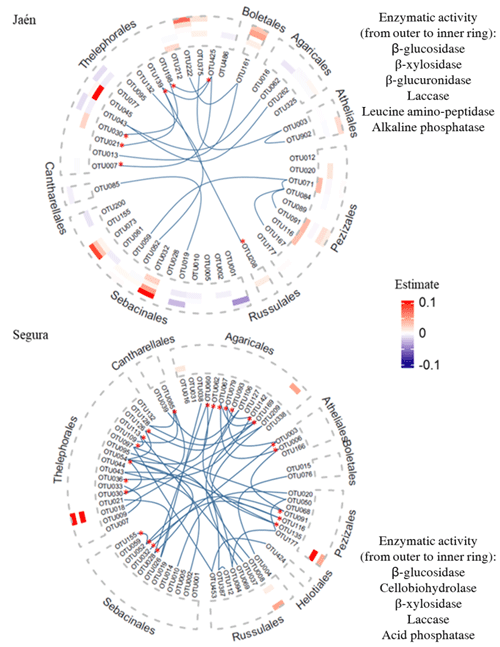
Figure 3Co-occurrence ECM fungal networks and contributions of fungal OTUs to soil extracellular enzymatic activity in Jaén and Segura. Co-occurring OTUs are marked with bold blue lines in the graphs and classed by fungal orders and keystone taxa with asterisks. Heatmaps show the estimated values (positive in red and negative in blue) of ECM fungal OTUs, explaining at least one soil enzymatic activity by elastic net (ENET) regularization models. The estimated values are 0.1-scaled. We used order as the main resolving taxonomic category to visualize trends of ECM fungi in predicting enzymatic activities within a network context and incorporating phylogenetic relationships. The ECM fungal OTU taxonomic assignment and estimated values are listed in Table S3.
In Segura, only six OTUs positively affected soil enzymatic activities (half of them either one or two enzymatic activities), particularly those related to C cycling (Fig. 3, Table S3b). The greater estimated values were detected for Peziza succosella and species belonging to Thelephoraceae, which positively explained β-glucosidase and β-xylosidase activities (Fig. 3, Table S3b).
ECM fungal co-occurrence networks in Mediterranean forest soils reveal a strong habitat dependency, supporting our first hypothesis. Network metrics differ between the two study forests by season and also by the host plant identity (i.e., Cistus vs. Quercus spp.) in Segura. When investigating the ECM fungal co-occurring network properties further, the abundance of occurring taxa appears to be a key parameter for network structuring since, for a given ECM fungal OTU, centrality metrics diminish with increasing OTU abundance. The co-occurrence network structure does not have a significant effect on the soil multi-enzymatic activity MAI, but we do find relationships when functional network metrics are analyzed (e.g., OTUs and their co-occurrences explaining at least one soil enzymatic activity), partially supporting our second hypothesis. Our study reveals that fungal OTUs within a co-occurrence network context may explain soil enzymatic activities (mostly those related to carbon cycling) and that major effects are directed by taxa belonging to the orders Thelephorales, Sebacinales and Pezizales.
4.1 Ectomycorrhizal fungal networks differed between Mediterranean mixed forests
ECM fungal co-occurrence networks in the study on Mediterranean mixed forests showed a strong habitat dependency. Environmental factors such as summer drought, wildfire recurrence or soil nutrient retention have been identified as important long-term habitat filters affecting Mediterranean forest community adaptations (Pérez-Valera et al., 2017; Pérez-Izquierdo et al., 2020; Hernandez et al., 2021). These filters could contribute to the regional changes in fungal OTU richness and network parameters observed in this study. In addition, soil properties were related to the outcomes associated with co-occurrence network complexity. Co-occurrence networks revealed differential responses to soil pH ranging from 6.9 to 8.3 (averaging 7.7±0.1) and from 4.9 to 8.2 (averaging 6.6±0.2) in Jaén and Segura, respectively, suggesting local adaptation to homogeneous neutral–alkaline soils in Jaén, in contrast to those more variable in Segura, which also depended on the host plant species identity, e.g., higher soil pH under C. albidus than Quercus spp. (Prieto-Rubio et al., 2023). On the basis that pH is a well-known filter for soil microbial community assembly (Glassman et al., 2017; Tripathi et al., 2018), our results suggest that neutral pH contributed to diminished network complexity, showing an increased number of potential single-OTU modules (Jaén). By contrast, neutral–acidic conditions could imply a wider range of available niches for ECM fungi, which would explain the increasing fungal network complexity observed in Segura.
The seasonality effect on the network parameters varied in each study forest, suggesting that this effect is dependent on the habitat characteristics. This habitat dependency of seasonal outcomes associated with nutrient availability and fungal communities in forest soils has been shown previously (Siles and Margesin, 2017; Rincón et al., 2015; Ji et al., 2021). In fact, the altitudinal variations between Jaén and Segura (∼300 m) may imply variations in physical–chemical and SOM properties, as shown previously in other Mediterranean forest ecosystems (Gutiérrez-Girón et al., 2015; Centenaro et al., 2023). These altitudinal variations also span the limit between meso- and supra-Mediterranean bioclimatic belts (∼1200 m a.s.l.) and lead to changes in the physiological responses of host plant communities (Rivas-Martínez, 1987) and likely also in the ECM fungal co-occurrence networks. Further studies including a broader altitudinal zonation would be needed to confirm this causal effect on Mediterranean forest soil communities.
4.2 The dominance of certain ECM fungi was key in network structuring
The richness and abundance of ECM fungal taxa were key to understanding how co-occurrence networks were structured. As argued by Wagg et al. (2019), the network complexity detrended by OTU richness discards the intrinsic variation due to OTU richness. This parameter revealed a habitat effect: that is, in the low-complexity network of Jaén, detrending network metrics helped find fine-tuned community outcomes that would be obscured by the low OTU richness, e.g., detecting seasonal variations. By contrast, in the more complex network of Segura, those seasonal variations were not detected but showed a host plant effect differing at the genus level (Cistus vs. Quercus) (as previously observed when phylogenetic turnover outcomes of the ECM fungal communities were evaluated, Prieto-Rubio et al., 2022). The network composition also varied between the studied forests but revealed common fungal orders (Pezizales, Sebacinales and Thelephorales) that dominated the network in Jaén (∼80 % of the total OTUs), whilst others (e.g., Agaricales, Boletales or Russulales) rarely occurred. In Segura, the abundance of the common fungal orders found in Jaén decreased, allowing other fungal lineages to increase their network representativeness (e.g., OTUs belonging to Agaricales and Atheliales). This contrasting pattern among fungal lineages might suggest that ECM fungal communities conserved a regional species pool with lineages likely differing in their ecological niche space, as observed in arbuscular mycorrhizal fungal communities by Davison et al. (2021).
The negative correlation found between fungal abundance and centrality metrics suggests that local dominance plays a key role in ECM fungal network structuring (Lamanna et al., 2016; Prieto-Rubio et al., 2022). This property of ECM fungi is consistent with that observed in arbuscular mycorrhizal fungal communities (Davison et al., 2022), and its extent could also be influenced by the environmental context (Wang et al., 2022). In Jaén, the edaphic environment (neutral–alkaline soils with high organic matter content and homogeneous under all host plant species, Prieto-Rubio et al., 2023) could limit niche breadth for ECM fungi, likely leaving a more important role to dominance and explaining the observed lower community richness and network complexity (Davison et al., 2022), whilst in Segura, where soil properties were more differentiated across host plant species (Prieto-Rubio et al., 2022, 2023), the increasing niche breadth for ECM fungi could mitigate the role of dominance (Davison et al., 2022). This would facilitate the coexistence among more fungal taxa, hence explaining the higher observed community richness and network complexity in this forest (Chomicki et al., 2019). Moreover, the outcomes concerning keystone taxa could also be affected by dominance, as revealing that these OTUs were not locally abundant confirmed that other mechanisms besides competitive exclusion, such as those related to demographic stochasticity (e.g., priority effects), could impact the co-occurrence network structure (Kennedy et al., 2009; Debray et al., 2022). Further studies decoupling the role of niche processes from that of stochasticity in interaction or co-occurrence networks will bring novel understandings of fungal community assembly.
4.3 ECM fungal network structure predicted enzymatic activities in forest soils
Contrary to our expectations, the overall network properties barely explained the soil MAI. However, when functional network properties inferred from ENET regularization were tested, we found that an increase in the number of ECM fungal OTUs related positively to any soil enzymatic activity, leading to positive soil MAI values. These patterns were mainly observed in Jaén, where a greater number of ECM fungal taxa explained any soil enzymatic activity compared with those in Segura. The effects of ECM fungal networks on enzymatic activities configuring the soil MAI were also visualized through the co-occurrence networks. Across the study forests, different ECM fungal lineages could be linked to the enzymatic activities, as previously observed in soil microbial communities from different ecotones (Trivedi et al., 2016; Banerjee et al., 2018a). The number of ECM fungal OTUs explaining soil enzymatic activities substantially differed between the study forests, with a greater number of OTUs explaining enzymatic activities in Jaén than in Segura. In accordance with microbial networks in grasslands (Wagg et al., 2019; Luo et al., 2022), we observed that highly complex ECM fungal networks presented fewer taxa explaining enzymatic activity rates in forest soils, i.e., as if increasing network complexity would lead to functional redundancy, likely explained by the functional complementarity among OTUs conforming to the network (Wang et al., 2021). This result supporting the ideas of functional redundancy (i.e., enzymatic activity conservation across ECM fungi, Baldrian and Kohout, 2017) fits well into the holobiont theory (Zobel et al., 2024). Indeed, from the holobiont perspective, this redundancy ensures that essential functions carried out by microbiota are maintained even if one or more species are lost or disrupted, providing resilience against environmental changes, disturbances or the loss of specific microbial species (Vandenkoornhuyse et al., 2015). Alternatively, other soil microbial groups could be contributing to P and N mobilization to a much greater extent than ECM in these forest soils (Ward et al., 2021; Xun et al., 2021).
Furthermore, we found that the effects of ECM fungal taxa on soil enzymatic activities varied across ECM fungal lineages. In Jaén, positive or negative effects on soil enzymatic activities related to C mobilization were shown in taxa belonging to Agaricales, Atheliales, Cantharellales, Pezizales and Russulales. These ECM fungal lineages are known to conserve exoenzyme pools related to C cycling (Miyauchi et al., 2020). We could also show that OTUs belonging to Pezizales mainly drove P mobilization, whilst taxa contributing to explaining N cycling were few but phylogenetically widespread across ECM fungal lineages. Our results also show that keystone ECM fungal taxa were not major determinants of soil enzymatic activities, especially in Segura, where few effects were detected despite the higher richness and network complexity. This suggests that keystone ECM fungal taxa play community assembly-related roles rather than those associated with biogeochemical functions, as initially assumed for these microbial groups in soil networks (Banerjee et al., 2018b). These findings show the need in further studies to integrate other microbial guilds (e.g., saprotrophic bacteria and fungi) into co-occurrence networks to deepen the soil community attributes that explain carbon and nutrient mobilization.
This study shows the importance of integrating co-occurrence network analyses to disentangle ectomycorrhizal fungal community structure and function. We conclude that ECM co-occurrence network outcomes are highly dependent on habitat characteristics and that increased network complexity could imply functional redundancy in forest soils. The abundance of ECM fungal taxa is key to understanding the network structure, supporting the potential role of dominance in the ECM fungal community assembly in Mediterranean forests. In this co-occurrence network context, we found that several ECM fungal lineages explained soil functioning through enzymatic activities. In addition, keystone ECM fungal taxa were not necessarily those that profoundly explain soil enzymatic activities, showing a main influence on the network structuring. Incorporating other soil microbial guilds (e.g., saprotrophs, pathogens or arbuscular mycorrhizas) into co-occurrence network analyses will help to accurately discern structural and biogeochemical roles within forest soil communities.
Datasets (information in Prieto-Rubio et al., 2022, and Prieto-Rubio et al., 2023) and R codes are available upon request.
The supplement related to this article is available online at: https://doi.org/10.5194/soil-10-425-2024-supplement.
JMA, CAA and AR designed the research. JPR, JLG, JMA, CAA and AR performed the sampling. JPR did the laboratory work. JPR, ALG and AR analyzed the data, and JPR wrote the manuscript. ALG and AR contributed equally. All the authors revised the manuscript.
The contact author has declared that none of the authors has any competing interests.
Publisher’s note: Copernicus Publications remains neutral with regard to jurisdictional claims made in the text, published maps, institutional affiliations, or any other geographical representation in this paper. While Copernicus Publications makes every effort to include appropriate place names, the final responsibility lies with the authors.
We acknowledge Leticia Pérez-Izquierdo and Lise Pomàrede for their help with samplings in the Natural Parks and Lola López for her technical support.
This research has been supported by the Agencia Estatal de Investigación (grant no. CGL2015-69118-C2-2P) and a Plan 6-UJA postdoctoral fellowship from the University of Jaén (Spain).
The article processing charges for this open-access publication were covered by the CSIC Open Access Publication Support Initiative through its Unit of Information Resources for Research (URICI).
This paper was edited by Lisa Ciadamidaro and reviewed by César Marín and one anonymous referee.
Abarenkov, K., Nilsson, R. H., Larsson, K. H., Alexander, I. J., Eberhardt, U., Erland, S., Høiland, K., Kjøller, R., Larsson, E., Pennanen, T., Sen, R., Taylor, A. F. S., Tedersoo, L., Ursing, B. M., Vrålstad, T., Liimatainen, K., Peintner, U., and Kõljalg, U.: The UNITE database for molecular identification of fungi-recent updates and future perspectives, New Phytol., 186, 281–285, https://doi.org/10.1111/j.1469-8137.2009.03160.x, 2010.
Alcántara, J. M., Pulgar, M., Trøjelsgaard, K., Garrido, J. L., and Rey, P. J.: Stochastic and deterministic effects on interactions between canopy and recruiting species in forest communities, Funct. Ecol., 32, 2264–2274, https://doi.org/10.1111/1365-2435.13140, 2018.
Azcón-Aguilar, C. and Barea, J. M.: Nutrient cycling in the mycorrhizosphere, J. Soil Sci. Plant Nut., 15, 372–396. https://doi.org/10.4067/s0718-95162015005000035, 2015.
Bahram, M., Netherway, T., Hildebrand, F., Pritsch, K., Drenkhan, R., Loit, K., Anslan, S., Bork, P., and Tedersoo, L.: Plant nutrient-acquisition strategies drive topsoil microbiome structure and function, New Phytol., 227, 1189–1199, https://doi.org/10.1111/nph.16598, 2020.
Banerjee, S., Thrall, P. H., Bissett, A., van der Heijden, M. G. A., and Richardson, A. E.: Linking microbial co-occurrences to soil ecological processes across a woodland-grassland ecotone, Ecol. Evol., 17, 8217–8230, https://doi.org/10.1002/ece3.4346, 2018a.
Banerjee, S., Schlaeppi, K., and van der Heijden, M. G. A.: Keystone taxa as drivers of microbiome structure and functioning, Nat. Rev. Microbiol., 16, 567–576, https://doi.org/10.1038/s41579-018-0024-1, 2018b.
Bates, D., Maechler, M., Bolker, B., and Walker, S.: Fitting Linear Mixed-Effects Models Using lme4, J. Stat. Soft., 67, 1–48, https://doi.org/10.18637/jss.v067.i01, 2015.
Barberán, A., Bates, S. T., Casamayor, E. O., and Fierer, N.: Using network analysis to explore co-occurrence patterns in soil microbial communities, ISME J., 6, 343–351, https://doi.org/10.1038/ismej.2011.119, 2012.
Barner, A. K., Coblentz, K. E., Hacker, S. D., and Menge, B. A.: Fundamental contradictions among observational and experimental estimates of non-trophic species interactions, Ecology, 99, 557–566. https://doi.org/10.1002/ecy.2133, 2018.
Barroso-Bergadà, D., Pauvert, C., Vallance, J., Delière, L., Bohan, D. A., Buée, M., and Vacher, C.: Microbial networks inferred from environmental DNA data for biomonitoring ecosystem change: Strengths and pitfalls, Mol. Ecol. Resour., 21, 1–19, https://doi.org/10.1111/1755-0998.13302, 2020.
Baldrian, P. and Kohout, P.: Interactions of saprotrophic fungi with tree roots: can we observe the emergence of novel ectomycorrhizal fungi?, New Phytol., 215, 511–513, https://doi.org/10.1111/nph.14665, 2017.
Bastian, M., Heymann, S., and Jacomy, M.: Gephi: an open source software for exploring and manipulating networks, in: Proceedings of the international AAAI conference on web and social media, 3, 361–362, https://doi.org/10.1609/icwsm.v3i1.13937, 2009.
Bastida, F., García, C., Fierer, N., Eldridge, D. J., Bowker, M. A., Abades, S., Alfaro, F. D., Behre, A. A., Cutler, N. A., Gallardo, A., García-Velázquez, L., Hart, S. C., Hayes, P. E., Hernández, T., Hseu, Z., Jehmlich, M., Kirchmair, M., Lambers, H., Neuhauser, S., Peña-Ramírez, V. M., Pérez, C. A., Reed, S. C., Santos, F., Siebe, C., Sullivan, B. W., Trivedi, P., Vera, A., Williams, M. A., Moreno, J. L., and Delgado-Baquerizo, M.: Global ecological predictors of the soil priming effect, Nat. Commun., 10, 1–9, https://doi.org/10.1038/s41467-019-11472-7, 2019.
Berry, D. and Widder, S.: Deciphering microbial interactions and detecting keystone species with co-occurrence networks, Front. Microbiol., 5, 90985, https://doi.org/10.3389/fmicb.2014.00219, 2014.
Blondel, V. D., Guillaume, J. L., Lambiotte, R., and Lefebvre, E.: Fast unfolding of communities in large networks, J. Stat. Mech.-Theory E., 2008, P10008, https://doi.org/10.1088/1742-5468/2008/10/P10008, 2008.
Carvalhais, L. C. and Dennis, P. G. (Eds): The Plant Microbiome: Methods and Protocols, Springer US, 155–171, https://doi.org/10.1007/978-1-0716-1040-4, 2021.
Centenaro, G., de Miguel, S., Amouzgar, L., Piñuela, Y., Son, D., Bonet, J. A., Aragon, J. M., Dashevskaya, S., Cataño, C., and Alday, J. G.: Silvicultural management and altitude prevail on soil properties and fungal community in shaping understorey plant communities in a Mediterranean pine forest, Sci. Total Environ., 858, 159860, https://doi.org/10.1016/j.scitotenv.2022.159860, 2023.
Cheeke, T. E., Phillips, R. P., Brzostek, E. R., Rosling, A., Bever, J. D., and Fransson, P.: Dominant mycorrhizal association of trees alters carbon and nutrient cycling by selecting for microbial groups with distinct enzyme function, New Phytol., 214, 432–442, https://doi.org/10.1111/nph.14343, 2017.
Chomicki, G., Weber, M., Antonelli, A., Bascompte, J., and Kiers, E. T.: The impact of mutualisms on species richness, Trends Ecol. Evol., 34, 698–711, https://doi.org/10.1016/j.tree.2019.03.003, 2019.
Cole, J. R., Wang, Q., Fish, J. A., Chai, B., McGarrell, D. M., Sun, Y., Brown, C. T., Porras-Alfaro, A., Kuske, C. R., and Tiedje, J.M.: Ribosomal Database Project: Data and tools for high throughput rRNA analysis, Nucl. Acid. Res., 42, 633–642, https://doi.org/10.1093/nar/gkt1244, 2014.
Courty, P. E., Buée, M., Diedhiou, A. G., Frey-Klett, P., Le Tacon, F., Rineau, F., Turpault, M. P., Uroz, S., and Garbaye, J.: The role of ectomycorrhizal communities in forest ecosystem processes: New perspectives and emerging concepts, Soil Biol. Biochem., 42, 679–698, https://doi.org/10.1016/j.soilbio.2009.12.006, 2010.
Csardi, G. and Nepusz, T.: The igraph software package for complex network research, InterJournal, complex systems, 1695, 1–9, https://igraph.org (last access: 31 July 2023), 2006.
Davison, J., Moora, M., Semchenko, M., Adenan, S. B., Ahmed, T., Akhmetzhanova, A. A., Alatalo, J. M., Al-Quraishy, S., Andriyanova, E., Anslan, S., Bahram, M., Batbaatar, A., Brown, C., Bueno, C. G., Cahill, J., Cantero, J. J., Casper, B. B., Cherosov, M., Chideh, S., Coelho, A. P., Coghill, M., Decocq, G., Dudov, S., Chimbioputo Fabiano, E., Fedosov, V. E., Fraser, L., Glassman, S. I., Helm, A., Henry, H. A. L., Hérault, B., Hiiesalu, I., Hiiesalu, I., Hozzein, W. N., Kohout, P., Kõljalg, U., Koorem, K., Laanisto, L., Mander, Ü., Mucina, L., Munyampundu, J., Neuenkamp, L., Niinemets, Ü., Nyamukondiwa, C., Oja, J., Onipchenko, V., Pärtel, M., Phosri, C., Põlme, S., Püssa, K., Ronk, A., Saitta, A., Semboli, O., Sepp, S., Seregin, A., Sudheer, S., Peña-Venegas, C. P., Paz, C., Vahter, T., Vasar, M., Veraart, A. J., Tedersoo, L., Zobel, M., and Öpik, M.: Temperature and pH define the realised niche space of arbuscular mycorrhizal fungi, New Phytol., 231, 763–776, https://doi.org/10.1111/nph.17240, 2021.
Davison, J., Vasar, M., Sepp, S. K., Oja, J., Al-Quraishy, S., Bueno, C. G., Cantero, J. J., Fabiano, E. C., Decocq, G., Fraser, L., Hiiesalu, I., Hozzein, W. N., Koorem, K., Moora, M., Mucina, L., Onipchenko, V., Öpik, M., Pärtel, M., Phosri, C., Semchenko, M., Vahter, T., Tedersoo, L., and Zobel, M.: Dominance, diversity, and niche breadth in arbuscular mycorrhizal fungal communities, Ecology, 103, e3761, https://doi.org/10.1002/ecy.3761, 2022.
Debray, R., Herbert, R. A., Jaffe, A. L., Crits-Christoph, A., Power, M. E., and Koskella, B.: Priority effects in microbiome assembly, Nat. Rev. Microbiol., 20, 109–121, https://doi.org/10.1038/s41579-021-00604-w, 2022.
Deguchi, T., Takahashi, K., Takayasu, H., and Takayasu, M.: Hubs and authorities in the world trade network using a weighted HITS algorithm, PLoS ONE, 9, 1–16, https://doi.org/10.1371/journal.pone.0100338, 2014.
Delgado-Baquerizo, M., Maestre, F. T., Reich, P. B., Jeffries, T. C., Gaitan, J. J., Encinar, D., Berdugo, M., Campbell, C. D., and Singh, B. K.: Microbial diversity drives multifunctionality in terrestrial ecosystems, Nat. Commun., 7, 1–8, https://doi.org/10.1038/ncomms10541, 2016.
Delmas, E., Besson, M., Brice, M. H., Burkle, L. A., dalla Riva, G. V., Fortin, M. J., Gravel, D., Guimarães Jr, P. R., Hembry, D. H., Newman, E. A., Olesen, J. M., Pires, M. M., Yeakel, J. D., and Poisot, T.: Analysing ecological networks of species interactions, Biol. Rev., 94, 16–36, https://doi.org/10.1111/brv.12433, 2019.
Fruchterman, T. M. and Reingold, E. M.: Graph drawing by force-directed placement, Softw. Pract. Exper., 21, 1129–1164, https://doi.org/10.1002/spe.4380211102, 1991.
Garrido, J. L., Alcántara, J. M., López-García, Á., Ozuna, C. V., Perea, A. J., Prieto, J., Rincón, A., and Azcón-Aguilar, C.: The structure and ecological function of the interactions between plants and arbuscular mycorrhizal fungi through multilayer networks, Funct. Ecol., 37, 2217–2230, https://doi.org/10.1111/1365-2435.14378, 2023.
Glassman, S. I., Wang, I. J., and Bruns, T. D.: Environmental filtering by pH and soil nutrients drives community assembly in fungi at fine spatial scales, Mol. Ecol., 26, 6960–6973, https://doi.org/10.1111/mec.14414, 2017.
Gloor, G. B., Macklaim, J. M., Pawlowsky-Glahn, V., and Egozcue, J. J.: Microbiome datasets are compositional: And this is not optional, Front. Microbiol., 8, 2224, https://doi.org/10.3389/fmicb.2017.02224, 2017.
Goberna, M. and Verdú, M.: Cautionary notes on the use of co-occurrence networks in soil ecology, Soil Biol. Biochem., 166, 108534, https://doi.org/10.1016/j.soilbio.2021.108534, 2022.
Götzenberger, L., de Bello, F., Bråthen, K. A., Davison, J., Dubuis, A., Guisan, A., Lepš, J., Lindborg, R., Moora, M., Pärtel, M., Pellissier, L., Pottier, J., Vittoz, P., Zobel, K., and Zobel, M.: Ecological assembly rules in plant communities-approaches, patterns and prospects, Biol. Rev., 87, 111–127, https://doi.org/10.1111/j.1469-185X.2011.00187.x, 2012.
Gouveia, C., Móréh, Á., and Jordán, F.: Combining centrality indices: Maximizing the predictability of keystone species in food webs, Ecol. Indic., 126, 107617, https://doi.org/10.1016/j.ecolind.2021.107617, 2021.
Gutiérrez-Girón, A., Díaz-Pinés, E., Rubio, A., and Gavilán, R. G.: Both altitude and vegetation affect temperature sensitivity of soil organic matter decomposition in Mediterranean high mountain soils, Geoderma, 237, 1–8, https://doi.org/10.1016/j.geoderma.2014.08.005, 2015.
Hernandez, D. J., David, A. S., Menges, E. S., Searcy, C. A., and Afkhami, M. E.: Environmental stress destabilizes microbial networks, ISME J., 15, 1722–1734, https://doi.org/10.1038/s41396-020-00882-x, 2021.
Jacomy, M., Venturini, T., Heymann, S., and Bastian, M.: ForceAtlas2, a continuous graph layout algorithm for handy network visualization designed for the Gephi software, PLoS ONE, 9, 1–12, https://doi.org/10.1371/journal.pone.0098679, 2014.
Ji, L., Yang, Y., and Yang, L.: Seasonal variations in soil fungal communities and co-occurrence networks along an altitudinal gradient in the cold temperate zone of China: A case study on Oakley Mountain, Catena, 204, 105448, https://doi.org/10.1016/j.catena.2021.105448, 2021.
Kadowaki, K., Yamamoto, S., Sato, H., Tanabe, A. S., Hidaka, A., and Toju, H.: Mycorrhizal fungi mediate the direction and strength of plant–soil feedbacks differently between arbuscular mycorrhizal and ectomycorrhizal communities, Commun. Biol., 1, 196, https://doi.org/10.1038/s42003-018-0201-9, 2018.
Kennedy, P. G., Peay, K. G., and Bruns, T. D.: Root tip competition among ectomycorrhizal fungi: Are priority effects a rule or an exception?, Ecology, 90, 2098–2107, https://doi.org/10.1890/08-1291.1, 2009.
Krapu, C. and Borsuk, M.: A spatial community regression approach to exploratory analysis of ecological data, Methods Ecol. Evol., 11, 608–620, https://doi.org/10.1111/2041-210X.13371, 2020.
Kurtz, Z. D., Müller, C. L., Miraldi, E. R., and Littman, D. R.: Sparse and Compositionally Robust Inference of Microbial Ecological Networks, Plos Comput. Biol., 11, 1–25, https://doi.org/10.1371/journal.pcbi.1004226, 2015.
Lamanna, J. A., Walton, M. L., Turner, B. L., and Myers, J. A.: Negative density dependence is stronger in resource-rich environments and diversifies communities when stronger for common but not rare species, Ecol. Lett., 19, 657–667, https://doi.org/10.1111/ele.12603, 2016.
Lassalle, G., Credoz, A., Hédacq, R., Bertoni, G., Dubucq, D., Fabre, S., and Elger, A.: Estimating persistent oil contamination in tropical region using vegetation indices and random forest regression, Ecotox. Environ. Safe., 184, 109654, https://doi.org/10.1016/j.ecoenv.2019.109654, 2019.
Lebreton, A., Zeng, Q., Miyauchi, S., Kohler, A., Dai, Y. C., and Martin, F. M.: Evolution of the mode of nutrition in symbiotic and saprotrophic fungi in forest ecosystems, Annu. Rev. Ecol. Evol. S., 52, 385–404, https://doi.org/10.1146/annurev-ecolsys-012021-114902, 2021.
Le Bagousse-Pinguet, Y., Soliveres, S., Gross, N., Torices, R., Berdugo, M., and Maestre, F. T.: Phylogenetic, functional, and taxonomic richness have both positive and negative effects on ecosystem multifunctionality, P. Natl. Acad. Sci. USA, 116, 8419–8424, https://doi.org/10.1073/pnas.1815727116, 2019.
Liang, M., Johnson, D., Burslem, D. F. R. P., Yu, S., Fang, M., Taylor, J. D., Taylor, A. F. S., Helgason, T., and Liu, X.: Soil fungal networks maintain local dominance of ectomycorrhizal trees, Nat. Commun., 11, 1–7, https://doi.org/10.1038/s41467-020-16507-y, 2020.
Liu, H., Roeder, K., and Wasserman, L.: Stability Approach to Regularization Selection (StARS) for High Dimensional Graphical Models, Advances in Neural Information Processing Systems, 23, https://proceedings.neurips.cc/paper_files/paper/2010/file/301ad0e3bd5cb1627a2044908a42fdc2-Paper.pdf (last access: 18 June 2024), 2010.
Liu, S., García-Palacios, P., Tedersoo, L., Guirado, E., van der Heijden, M. G., Wagg, C., Chen, D., Wang, Q., Wang, J., Singh, B. K., and Delgado-Baquerizo, M.: Phylotype diversity within soil fungal functional groups drives ecosystem stability, Nat. Ecol. Evol., 6, 900–909, https://doi.org/10.1038/s41559-022-01756-5, 2022.
Luo, S., Png, K., Ostle, N. J., Zhou, H., Hou, X., Luo, C., Quinton, J. N., Schaffner, U., Sweeney, C., Wang, D., Wu, J., Wu, Y., and Bardgett, R. D.: Grassland degradation-induced declines in soil fungal complexity reduce fungal community stability and ecosystem multifunctionality, Soil Biol. Biochem., 176, 108865, https://doi.org/10.1016/j.soilbio.2022.108865, 2022.
Mamet, S. D., Redlick, E., Brabant, M., Lamb, E. G., Helgason, B. L., Stanley, K., and Siciliano, S. D.: Structural equation modeling of a winnowed soil microbiome identifies how invasive plants re-structure microbial networks, ISME J., 13, 1988–1996, https://doi.org/10.1038/s41396-019-0407-y, 2019.
Mandakovic, D., Rojas, C., Maldonado, J., Latorre, M., Travisany, D., Delage, E., Bihouée, A., Jean, G., Díaz, F. P., Fernández-Gómez, B., Cabrera, P., Gaete, A., Latorre, C., Gutiérrez, R. A., Maass, A., Cambiazo, V., Navarrete, S. A., Eveillard, D., and González, M.: Structure and co-occurrence patterns in microbial communities under acute environmental stress reveal ecological factors fostering resilience, Sci. Rep., 8, 1–12, https://doi.org/10.1038/s41598-018-23931-0, 2018.
Martin, F. M. and van der Heijden, M. G. A.: The mycorrhizal symbiosis: research frontiers in genomics, ecology, and agricultural application, New Phytol., 242, 1486–1506, https://doi.org/10.1111/nph.19541, 2024.
Mazel, F., Pennell, M. W., Cadotte, M. W., Diaz, S., Dalla Riva, G. V., Grenyer, R., Leprieur, F., Mooers, A. O., Mouillot, D., Tucker, C. M., and Pearse, W.: Prioritizing phylogenetic diversity captures functional diversity unreliably, Nat Commun., 9, 2888, https://doi.org/10.1038/s41467-018-05126-3, 2018.
Meinshausen, N. and Bühlmann, P.: High-dimensional graphs and variable selection with the Lasso, Ann. Stat., 34, 1436–1462, https://doi.org/10.1214/009053606000000281, 2006.
Meyer, S. T., Ptacnik, R., Hillebrand, H., Bessler, H., Buchmann, N., Ebeling, A., Eisenhauer, N., Engels, C., Fischer, M., Halle, S., Klein, A., Oelmann, Y., Roscher, C., Rottstock, T., Scherber, C., Scheu, S., Schmid, B., Schulze, E., Temperton, V. M., Tscharntke, T., Voigt, W., Weigelt, A., Wilcke, W., and Weisser, W. W.: Biodiversity-multifunctionality relationships depend on identity and number of measured functions, Nat. Ecol. Evol., 2, 44–49, https://doi.org/10.1038/s41559-017-0391-4, 2018.
Miyauchi, S., Kiss, E., Kuo, A., Drula, E., Kohler, A., Sánchez-García, M., Morin, E., Andreopoulos, B., Barry, K. W., Bonito, G., Buée, M., Carver, A., Chen, C., Cichocki, N., Clum, A., Culley, D., Crous, P. W., Fauchery, L., Girlanda, M., Hayes, R. D., Kéri, Z., LaButti, K., Lipzen, A., Lombard, V., Magnuson, J., Maillard, F., Murat, C., Nolan, M., Ohm, R. A., Pangilinan, J., Pereira, M., Perotto, S., Peter, M., Pfister, S., Riley, R., Sitrit, Y., Stielow, J. B., Szöllõsi, G., Žifèáková, L., Štursová, M., Spatafora, J. W., Tedersoo, L., Vaario, L., Yamada, A., Yan, M., Wang, P., Xu, J., Bruns, T., Baldrian, P., Vilgalys, R., Dunand, C., Henrissat, B., Grigoriev, I. V., Hibbett, D., Nagy, L. G., and Martin, F. M.: Large-scale genome sequencing of mycorrhizal fungi provides insights into the early evolution of symbiotic traits, Nat. Commun., 11, 1–17, https://doi.org/10.1038/s41467-020-18795-w, 2020.
Morueta-Holme, N., Blonder, B., Sandel, B., McGill, B. J., Peet, R. K., Ott, J. E., Violle, C., Enquist, B. J., Jørgensen, P. M., and Svenning, J. C.: A network approach for inferring species associations from co-occurrence data, Ecography, 39, 1139–1150. https://doi.org/10.1111/ecog.01892, 2016.
Nguyen, D. Q., Schneider, D., Brinkmann, N., Song, B., Janz, D., Schöning, I., Daniel, R., and Polle, A.: Soil and root nutrient chemistry structure root-associated fungal assemblages in temperate forests, Environ. Microbiol., 22, 3081–3095, https://doi.org/10.1111/1462-2920.15037, 2020.
Nguyen, N. H.: Fungal Hyphosphere Microbiomes Are Distinct from Surrounding Substrates and Show Consistent Association Patterns, Microb. Spec., 11, e04708-22, https://doi.org/10.1128/spectrum.04708-22, 2023.
Nguyen, N. H., Song, Z., Bates, S. T., Branco, S., Tedersoo, L., Menke, J., Schilling, J. S., and Kennedy, P. G.: FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild, Fungal Ecol., 20, 241–248, https://doi.org/10.1016/j.funeco.2015.06.006, 2016.
Oksanen, J., Simpson, G. L., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P. R., O'Hara, R. B., Solymos, P., Stevens, M. H. H., Szoecs, E., Wagner, H., Barbour, M., Bedward, M., Bolker, B., Borcard, D., Carvalho, G., Chirico, M., De Caceres, M., Durand, S., Evangelista, H. B. A., FitzJohn, R., Friendly, M., Furneaux, B., Hannigan, G., Hill, M. O., Lahti, L., McGlinn, D., Ouellette, M., Cunha, E. D., Smith, T., Stier, A., Ter Braak, C. J. F., and Weedon, J.: vegan: Community Ecology Package, R package version 2.6-4, https://CRAN.R-project.org/package=vegan (last access: 30 September 2023), 2022.
Pajares-Murgó, M., Garrido, J. L., Perea, A. J., López-García, Á., and Alcántara, J. M.: Biotic filters driving the differentiation of decomposer, epiphytic and pathogenic phyllosphere fungi across plant species, Oikos, 2023, e09624, https://doi.org/10.1111/oik.09624, 2023.
Pauvert. C., Buée, M., Laval, V., Edel-Hermann, V., Fauchery, L., Gautier, A., Lesur, I., Vallance, J., and Vacher, C.: Bioinformatics matters: The accuracy of plant and soil fungal community data is highly dependent on the metabarcoding pipeline, Fungal Ecol., 41, 23–33, https://doi.org/10.1016/j.funeco.2019.03.005, 2019.
Pérez-Izquierdo, L., Zabal-Aguirre, M., Flores-Rentería, D., González-Martínez, S. C., Buée, M., and Rincón, A.: Functional outcomes of fungal community shifts driven by tree genotype and spatial-temporal factors in Mediterranean pine forests, Environ. Microbiol., 19, 1639–1652, https://doi.org/10.1111/1462-2920.13690, 2017.
Pérez-Izquierdo, L., Saint-André, L., Santenoise, P., Buée, M., and Rincón, A.: Tree genotype and seasonal effects on soil properties and biogeochemical functioning in Mediterranean pine forests, Eur. J. Soil Sci., 69, 1087–1097, https://doi.org/10.1111/ejss.12712, 2018.
Pérez-Izquierdo, L., Zabal-Aguirre, M., Verdú, M., Buée, M., and Rincón, A.: Ectomycorrhizal fungal diversity decreases in Mediterranean pine forests adapted to recurrent fires, Mol. Ecol., 1–14, 2463–2476, https://doi.org/10.1111/mec.15493, 2020.
Pérez-Valera, E., Goberna, M., Faust, K., Raes, J., García, C., and Verdú, M.: Fire modifies the phylogenetic structure of soil bacterial co-occurrence networks, Environ. Microbiol., 19, 317–327, https://doi.org/10.1111/1462-2920.13609, 2017.
Poudel, R., Jumpponen, A., Schlatter, D. C., Paulitz, T. C., Gardener, B. B. M., Kinkel, L. L., and Garrett, K. A.: Microbiome Networks: A Systems Framework for Identifying Candidate Microbial Assemblages for Disease Management, Phytopathology, 106, 1083–1096, https://doi.org/10.1094/PHYTO-02-16-0058-FI, 2016.
Pritsch, K. and Garbaye, J.: Enzyme secretion by ECM fungi and exploitation of mineral nutrients from soil organic matter, Ann. Forest Sci., 68, 25–32, https://doi.org/10.1007/s13595-010-0004-8, 2011.
Prieto-Rubio, J., Garrido, J. L., Pérez-Izquierdo, L., Alcántara, J. M., Azcón-Aguilar, C., López-García, A., and Rincón, A.: Scale dependency of ectomycorrhizal fungal community assembly processes in Mediterranean mixed forests, Mycorrhiza, 32, 315–325, https://doi.org/10.1007/s00572-022-01083-4, 2022.
Prieto-Rubio, J., Perea, A., Garrido, J. L., Alcántara, J. M., Azcón-Aguilar, C., López-García, A., and Rincón, A.: Plant Traits and Phylogeny Predict Soil Carbon and Nutrient Cycling in Mediterranean Mixed Forests, Ecosystems, 26, 1047–1060, https://doi.org/10.1007/s10021-022-00815-z, 2023.
Pulgar, M., Alcántara, J. M., and Rey, P. J.: Effects of sampling effort on estimates of the structure of replacement networks, J. Veg. Sci., 28, 445–457, https://doi.org/10.1111/jvs.12492, 2017.
Rincón, A., Santamaría, B. P., Ocaña, L., and Verdú, M.: Structure and phylogenetic diversity of post-fire ectomycorrhizal communities of maritime pine, Mycorrhiza, 24, 131–141, https://doi.org/10.1007/s00572-013-0520-0, 2014.
Rincón, A., Santamaría-Pérez, B., Rabasa, S. G., Coince, A., Marçais, B., and Buée, M.: Compartmentalized and contrasted response of ectomycorrhizal and soil fungal communities of s cots pine forests along elevation gradients in f rance and s pain, Environ. Microbiol., 17, 3009–3024, https://doi.org/10.1111/1462-2920.12894, 2015.
Rivas-Martínez, S.: Memoria del mapa de series de vegetación de España 1:400.000, 268 pp., ICONA, Ministerio de Agricultura, Pesca y Alimentación, Madrid, 1987.
Segnitz, R. M., Russo, S. E., Davies, S. J., and Peay, K. G.: Ectomycorrhizal fungi drive positive phylogenetic plant–soil feedbacks in a regionally dominant tropical plant family, Ecology, 101, 1–15, https://doi.org/10.1002/ecy.3083, 2020.
Siles, G., Rey, P. J., Alcántara, J. M., and Ramírez, J. M.: Assessing the long-term contribution of nurse plants to restoration of Mediterranean forests through Markovian models, J. App. Ecol., 45, 1790–1798, https://doi.org/10.1111/j.1365-2664.2008.01574.x, 2008.
Siles, J. A. and Margesin, R.: Seasonal soil microbial responses are limited to changes in functionality at two Alpine forest sites differing in altitude and vegetation, Sci. Rep., 7, 1–16, https://doi.org/10.1038/s41598-017-02363-2, 2017.
Smith, S. and Read, D.: Mycorrhizal Symbiosis, Elsevier Ltd., https://doi.org/10.1016/B978-0-12-370526-6.X5001-6, 2008.
Trivedi, P., Delgado-Baquerizo, M., Trivedi, C., Hu, H., Anderson, I. C., Jeffries, T. C., Zhou, J., and Singh, B. K.: Microbial regulation of the soil carbon cycle: Evidence from gene-enzyme relationships, ISME J., 10, 2593–2604, https://doi.org/10.1038/ismej.2016.65, 2008.
Trivedi, P., Delgado-Baquerizo, M., Trivedi, C., Hu, H., Anderson, I. C., Jeffries, T. C., Zou, J., and Singh, B. K.: Microbial regulation of the soil carbon cycle: evidence from gene-enzyme relationships, ISME J., 10, 2593–2604, https://doi.org/10.1038/ismej.2016.65, 2016.
Tripathi, B. M., Stegen, J. C., Kim, M., Dong, K., Adams, J. M., and Lee, Y. K.: Soil pH mediates the balance between stochastic and deterministic assembly of bacteria, ISME J., 12, 1072–1083, https://doi.org/10.1038/s41396-018-0082-4, 2018.
Van der Heijden, M. G., Martin, F. M., Selosse, M. A., and Sanders, I. R.: Mycorrhizal ecology and evolution: the past, the present, and the future, New Phytol., 205, 1406–1423, https://doi.org/10.1111/nph.13288, 2015.
Vandenkoornhuyse, P., Quaiser, A., Duhamel, M., Le Van, A., and Dufresne, A.: The importance of the microbiome of the plant holobiont, New Phytol., 206, 1196–1206, https://doi.org/10.1111/nph.13312, 2015.
Wagg, C., Schlaeppi, K., Banerjee, S., Kuramae, E. E., and van der Heijden, M. G.: Fungal-bacterial diversity and microbiome complexity predict ecosystem functioning, Nat. Commun., 10, 4841, https://doi.org/10.1038/s41467-019-12798-y, 2019.
Wagg, C., Hautier, Y., Pellkofer, S., Banerjee, S., Schmid, B., and van der Heijden, M. G. A.: Diversity and asynchrony in soil microbial communities stabilizes ecosystem functioning, ELife, 10, 1–19, https://doi.org/10.7554/eLife.62813, 2021.
Walkley, A. and Black, I. A.: An examination of the Degtjareff method for determining soil organic matter, and a proposed modification of the chromic acid titration method, Soil Sci., 37, 29–38, 1934.
Wang, J., Liao, L., Wang, G., Liu, H., Wu, Y., Liu, G., and Zhang, C.: N-induced root exudates mediate the rhizosphere fungal assembly and affect species coexistence, Sci. Total Environ., 804, 150148, https://doi.org/10.1016/j.scitotenv.2021.150148, 2022.
Wang, S., Isbell, F., Deng, W., Hong, P., Dee, L. E., Thompson, P., and Loreau, M.: How complementarity and selection affect the relationship between ecosystem functioning and stability, Ecology, 102, 1–12, https://doi.org/10.1002/ecy.3347, 2021.
Ward, E. B., Duguid, M. C., Kuebbing, S. E., Lendemer, J. C., Warren, R. J., and Bradford, M. A.: Ericoid mycorrhizal shrubs alter the relationship between tree mycorrhizal dominance and soil carbon and nitrogen, J. Ecol., 109, 3524–3540, https://doi.org/10.1111/1365-2745.13734, 2021.
Watts, S. C., Ritchie, S. C., Inouye, M., and Holt, K. E.: FastSpar: Rapid and scalable correlation estimation for compositional data, Bioinformatics, 35, 1064–1066, https://doi.org/10.1093/bioinformatics/bty734, 2019.
Wu, B.-W., Gao, C., Chen, L., Buscot, F., Goldmann, K., Purahong, W., Ji, N. N., Wang, Y. L., Lü, P. P., Li, X. C., and Guo, L. D.: Host Phylogeny Is a Major Determinant of Fagaceae-Associated Ectomycorrhizal Fungal Community Assembly at a Regional Scale, Front. Microbiol., 9, 1–12, https://doi.org/10.3389/fmicb.2018.02409, 2018.
Xun, W., Liu, Y., Li, W., Ren, Y., Xiong, W., Xu, Z., Zhang, N., Miao, Y., Shen, Q., and Zhang, R.: Specialized metabolic functions of keystone taxa sustain soil microbiome stability, Microbiome, 9, 1–15, https://doi.org/10.1186/s40168-020-00985-9, 2021.
Zhan, P., Liu, Y., Wang, H., Wang, C., Xia, M., Wang, N., Cui, W., Xiao, D., and Wang, H.: Plant litter decomposition in wetlands is closely associated with phyllospheric fungi as revealed by microbial community dynamics and co-occurrence network, Sci. Total Environ., 753, 142194, https://doi.org/10.1016/j.scitotenv.2020.142194, 2021.
Zobel, M., Koorem, K., Moora, M., Semchenko, M., and Davison, J.: Symbiont plasticity as a driver of plant success, New Phytol., 241, 2340–2352, https://doi.org/10.1111/nph.19566, 2024.
Zou, H. and Hastie, T.: Regularization and variable selection via the elastic net, J. Roy. Stat. Soc. B., 67, 301–320, https://doi.org/10.1111/j.1467-9868.2005.00503.x, 2005.